Benoit M. Dawant
IntraStyler: Exemplar-based Style Synthesis for Cross-modality Domain Adaptation
Jan 01, 2026Abstract:Image-level domain alignment is the de facto approach for unsupervised domain adaptation, where unpaired image translation is used to minimize the domain gap. Prior studies mainly focus on the domain shift between the source and target domains, whereas the intra-domain variability remains under-explored. To address the latter, an effective strategy is to diversify the styles of the synthetic target domain data during image translation. However, previous methods typically require intra-domain variations to be pre-specified for style synthesis, which may be impractical. In this paper, we propose an exemplar-based style synthesis method named IntraStyler, which can capture diverse intra-domain styles without any prior knowledge. Specifically, IntraStyler uses an exemplar image to guide the style synthesis such that the output style matches the exemplar style. To extract the style-only features, we introduce a style encoder to learn styles discriminatively based on contrastive learning. We evaluate the proposed method on the largest public dataset for cross-modality domain adaptation, CrossMoDA 2023. Our experiments show the efficacy of our method in controllable style synthesis and the benefits of diverse synthetic data for downstream segmentation. Code is available at https://github.com/han-liu/IntraStyler.
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
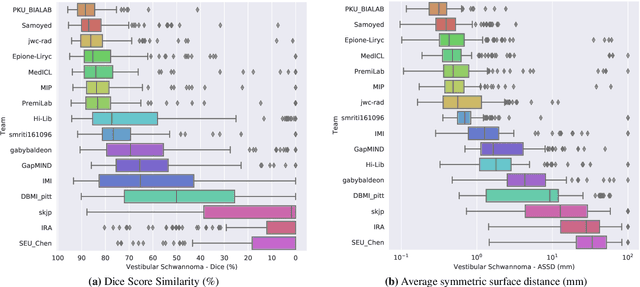
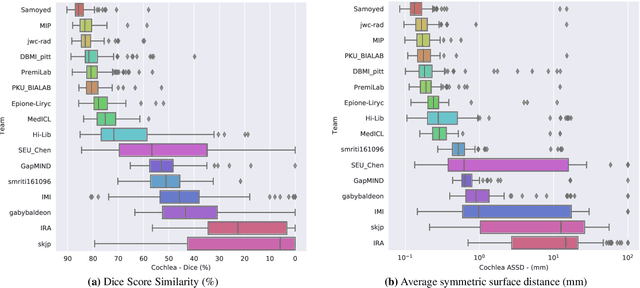
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
Learning Site-specific Styles for Multi-institutional Unsupervised Cross-modality Domain Adaptation
Nov 22, 2023



Abstract:Unsupervised cross-modality domain adaptation is a challenging task in medical image analysis, and it becomes more challenging when source and target domain data are collected from multiple institutions. In this paper, we present our solution to tackle the multi-institutional unsupervised domain adaptation for the crossMoDA 2023 challenge. First, we perform unpaired image translation to translate the source domain images to the target domain, where we design a dynamic network to generate synthetic target domain images with controllable, site-specific styles. Afterwards, we train a segmentation model using the synthetic images and further reduce the domain gap by self-training. Our solution achieved the 1st place during both the validation and testing phases of the challenge. The code repository is publicly available at https://github.com/MedICL-VU/crossmoda2023.
Evaluation of Synthetically Generated CT for use in Transcranial Focused Ultrasound Procedures
Oct 26, 2022



Abstract:Transcranial focused ultrasound (tFUS) is a therapeutic ultrasound method that focuses sound through the skull to a small region noninvasively and often under MRI guidance. CT imaging is used to estimate the acoustic properties that vary between individual skulls to enable effective focusing during tFUS procedures, exposing patients to potentially harmful radiation. A method to estimate acoustic parameters in the skull without the need for CT would be desirable. Here, we synthesized CT images from routinely acquired T1-weighted MRI by using a 3D patch-based conditional generative adversarial network (cGAN) and evaluated the performance of synthesized CT (sCT) images for treatment planning with tFUS. We compared the performance of sCT to real CT (rCT) images for tFUS planning using Kranion and simulations using the acoustic toolbox, k-Wave. Simulations were performed for 3 tFUS scenarios: 1) no aberration correction, 2) correction with phases calculated from Kranion, and 3) phase shifts calculated from time-reversal. From Kranion, skull density ratio, skull thickness, and number of active elements between rCT and sCT had Pearson's Correlation Coefficients of 0.94, 0.92, and 0.98, respectively. Among 20 targets, differences in simulated peak pressure between rCT and sCT were largest without phase correction (12.4$\pm$8.1%) and smallest with Kranion phases (7.3$\pm$6.0%). The distance between peak focal locations between rCT and sCT was less than 1.3 mm for all simulation cases. Real and synthetically generated skulls had comparable image similarity, skull measurements, and acoustic simulation metrics. Our work demonstrates the feasibility of replacing real CTs with the MR-synthesized CT for tFUS planning. Source code and a docker image with the trained model are available at https://github.com/han-liu/SynCT_TcMRgFUS
Enhancing Data Diversity for Self-training Based Unsupervised Cross-modality Vestibular Schwannoma and Cochlea Segmentation
Sep 23, 2022


Abstract:Automatic segmentation of vestibular schwannoma (VS) and the cochlea from magnetic resonance imaging (MRI) can facilitate VS treatment planning. Unsupervised segmentation methods have shown promising results without requiring the time-consuming and laborious manual labeling process. In this paper, we present an approach for VS and cochlea segmentation in an unsupervised domain adaptation setting. Specifically, we first develop a cross-site cross-modality unpaired image translation strategy to enrich the diversity of the synthesized data. Then, we devise a rule-based offline augmentation technique to further minimize the domain gap. Lastly, we adopt a self-configuring segmentation framework empowered by self-training to obtain the final results. On the CrossMoDA 2022 validation leaderboard, our method has achieved competitive VS and cochlea segmentation performance with mean dice scores of 0.8178 $\pm$ 0.0803 and 0.8433 $\pm$ 0.0293, respectively.
Synthetic CT Skull Generation for Transcranial MR Imaging-Guided Focused Ultrasound Interventions with Conditional Adversarial Networks
Feb 22, 2022



Abstract:Transcranial MRI-guided focused ultrasound (TcMRgFUS) is a therapeutic ultrasound method that focuses sound through the skull to a small region noninvasively under MRI guidance. It is clinically approved to thermally ablate regions of the thalamus and is being explored for other therapies, such as blood brain barrier opening and neuromodulation. To accurately target ultrasound through the skull, the transmitted waves must constructively interfere at the target region. However, heterogeneity of the sound speed, density, and ultrasound attenuation in different individuals' skulls requires patient-specific estimates of these parameters for optimal treatment planning. CT imaging is currently the gold standard for estimating acoustic properties of an individual skull during clinical procedures, but CT imaging exposes patients to radiation and increases the overall number of imaging procedures required for therapy. A method to estimate acoustic parameters in the skull without the need for CT would be desirable. Here, we synthesized CT images from routinely acquired T1-weighted MRI by using a 3D patch-based conditional generative adversarial network and evaluated the performance of synthesized CT images for treatment planning with transcranial focused ultrasound. We compared the performance of synthetic CT to real CT images using Kranion and k-Wave acoustic simulation. Our work demonstrates the feasibility of replacing real CT with the MR-synthesized CT for TcMRgFUS planning.
A Multi-rater Comparative Study of Automatic Target Localization Methods for Epilepsy Deep Brain Stimulation Procedures
Jan 26, 2022Abstract:Epilepsy is the fourth most common neurological disorder and affects people of all ages worldwide. Deep Brain Stimulation (DBS) has emerged as an alternative treatment option when anti-epileptic drugs or resective surgery cannot lead to satisfactory outcomes. To facilitate the planning of the procedure and for its standardization, it is desirable to develop an algorithm to automatically localize the DBS stimulation target, i.e., Anterior Nucleus of Thalamus (ANT), which is a challenging target to plan. In this work, we perform an extensive comparative study by benchmarking various localization methods for ANT-DBS. Specifically, the methods involved in this study include traditional registration method and deep-learning-based methods including heatmap matching and differentiable spatial to numerical transform (DSNT). Our experimental results show that the deep-learning (DL)-based localization methods that are trained with pseudo labels can achieve a performance that is comparable to the inter-rater and intra-rater variability and that they are orders of magnitude faster than traditional methods.
Unsupervised Domain Adaptation for Vestibular Schwannoma and Cochlea Segmentation via Semi-supervised Learning and Label Fusion
Jan 25, 2022



Abstract:Automatic methods to segment the vestibular schwannoma (VS) tumors and the cochlea from magnetic resonance imaging (MRI) are critical to VS treatment planning. Although supervised methods have achieved satisfactory performance in VS segmentation, they require full annotations by experts, which is laborious and time-consuming. In this work, we aim to tackle the VS and cochlea segmentation problem in an unsupervised domain adaptation setting. Our proposed method leverages both the image-level domain alignment to minimize the domain divergence and semi-supervised training to further boost the performance. Furthermore, we propose to fuse the labels predicted from multiple models via noisy label correction. In the MICCAI 2021 crossMoDA challenge, our results on the final evaluation leaderboard showed that our proposed method has achieved promising segmentation performance with mean dice score of 79.9% and 82.5% and ASSD of 1.29 mm and 0.18 mm for VS tumor and cochlea, respectively. The cochlea ASSD achieved by our method has outperformed all other competing methods as well as the supervised nnU-Net.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022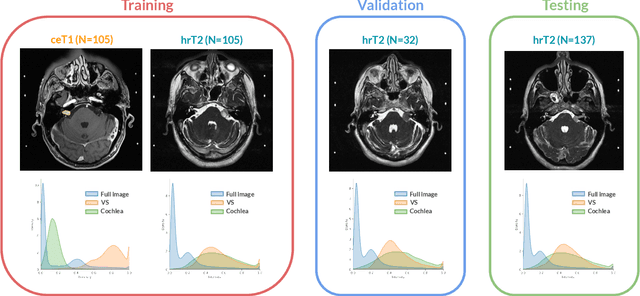
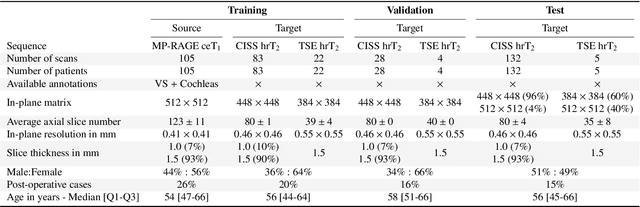
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
Cross-Modality Domain Adaptation for Vestibular Schwannoma and Cochlea Segmentation
Sep 21, 2021


Abstract:Automatic methods to segment the vestibular schwannoma (VS) tumors and the cochlea from magnetic resonance imaging (MRI) are critical to VS treatment planning. Although supervised methods have achieved satisfactory performance in VS segmentation, they require full annotations by experts, which is laborious and time-consuming. In this work, we aim to tackle the VS and cochlea segmentation problem in an unsupervised domain adaptation setting. Our proposed method leverages both the image-level domain alignment to minimize the domain divergence and semi-supervised training to further boost the performance. Furthermore, we propose to fuse the labels predicted from multiple models via noisy label correction. Our results on the challenge validation leaderboard showed that our unsupervised method has achieved promising VS and cochlea segmentation performance with mean dice score of 0.8261 $\pm$ 0.0416; The mean dice value for the tumor is 0.8302 $\pm$ 0.0772. This is comparable to the weakly-supervised based method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge