Willie Boag
Feature Robustness in Non-stationary Health Records: Caveats to Deployable Model Performance in Common Clinical Machine Learning Tasks
Aug 02, 2019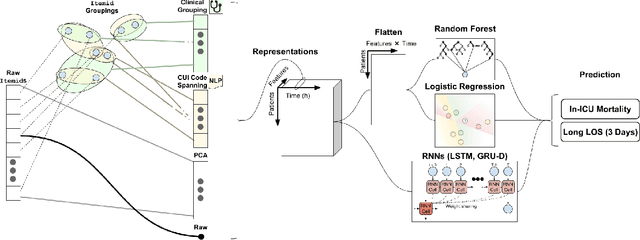
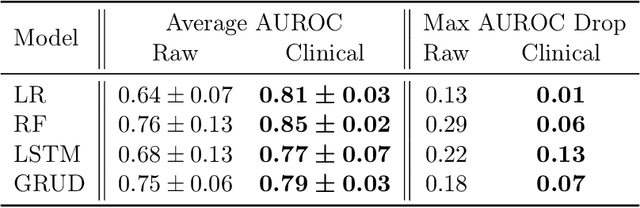
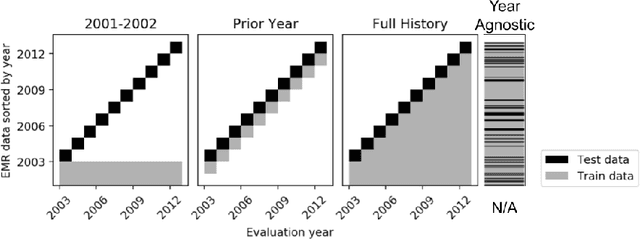
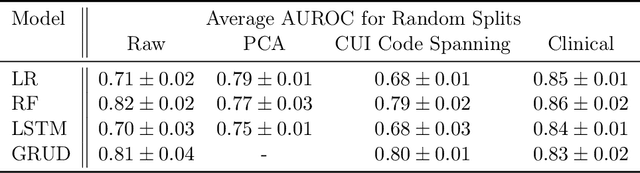
Abstract:When training clinical prediction models from electronic health records (EHRs), a key concern should be a model's ability to sustain performance over time when deployed, even as care practices, database systems, and population demographics evolve. Due to de-identification requirements, however, current experimental practices for public EHR benchmarks (such as the MIMIC-III critical care dataset) are time agnostic, assigning care records to train or test sets without regard for the actual dates of care. As a result, current benchmarks cannot assess how well models trained on one year generalise to another. In this work, we obtain a Limited Data Use Agreement to access year of care for each record in MIMIC and show that all tested state-of-the-art models decay in prediction quality when trained on historical data and tested on future data, particularly in response to a system-wide record-keeping change in 2008 (0.29 drop in AUROC for mortality prediction, 0.10 drop in AUROC for length-of-stay prediction with a random forest classifier). We further develop a simple yet effective mitigation strategy: by aggregating raw features into expert-defined clinical concepts, we see only a 0.06 drop in AUROC for mortality prediction and a 0.03 drop in AUROC for length-of-stay prediction. We demonstrate that this aggregation strategy outperforms other automatic feature preprocessing techniques aimed at increasing robustness to data drift. We release our aggregated representations and code to encourage more deployable clinical prediction models.
Publicly Available Clinical BERT Embeddings
Apr 29, 2019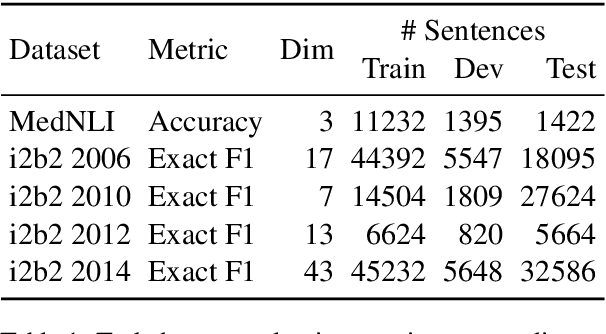
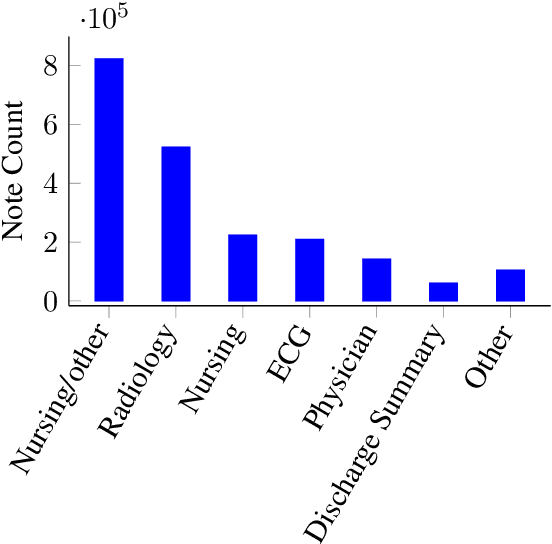
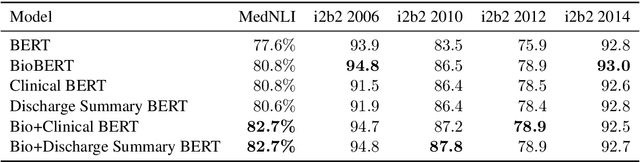
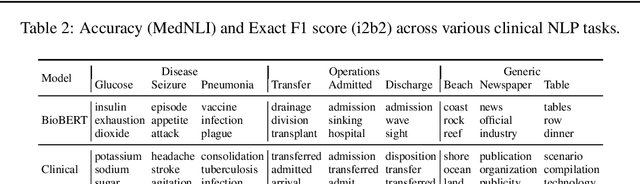
Abstract:Contextual word embedding models such as ELMo (Peters et al., 2018) and BERT (Devlin et al., 2018) have dramatically improved performance for many natural language processing (NLP) tasks in recent months. However, these models have been minimally explored on specialty corpora, such as clinical text; moreover, in the clinical domain, no publicly-available pre-trained BERT models yet exist. In this work, we address this need by exploring and releasing BERT models for clinical text: one for generic clinical text and another for discharge summaries specifically. We demonstrate that using a domain-specific model yields performance improvements on three common clinical NLP tasks as compared to nonspecific embeddings. These domain-specific models are not as performant on two clinical de-identification tasks, and argue that this is a natural consequence of the differences between de-identified source text and synthetically non de-identified task text.
Clinically Accurate Chest X-Ray Report Generation
Apr 04, 2019
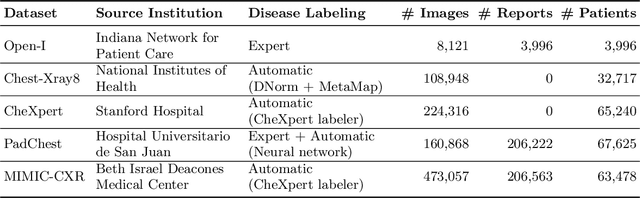
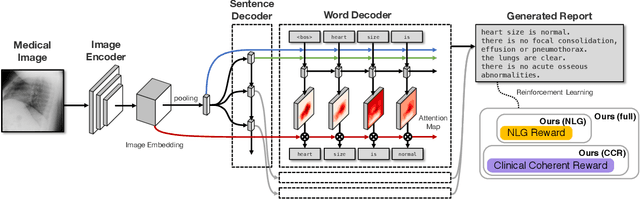
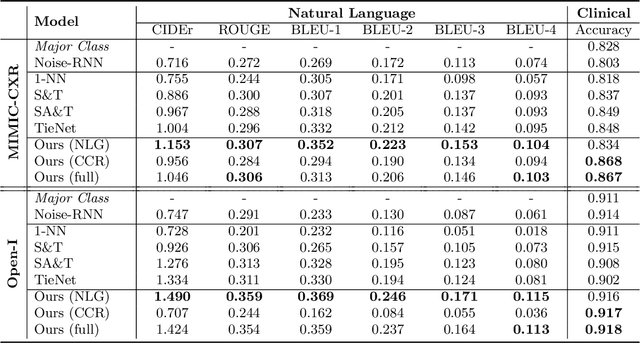
Abstract:The automatic generation of radiology reports given medical radiographs has significant potential to operationally and clinically improve patient care. A number of prior works have focused on this problem, employing advanced methods from computer vision and natural language generation to produce readable reports. However, these works often fail to account for the particular nuances of the radiology domain, and, in particular, the critical importance of clinical accuracy in the resulting generated reports. In this work, we present a domain-aware automatic chest X-Ray radiology report generation system which first predicts what topics will be discussed in the report, then conditionally generates sentences corresponding to these topics. The resulting system is fine-tuned using reinforcement learning, considering both readability and clinical accuracy, as assessed by the proposed Clinically Coherent Reward. We verify this system on two datasets, Open-I and MIMIC-CXR, and demonstrate that our model offers marked improvements on both language generation metrics and CheXpert assessed accuracy over a variety of competitive baselines.
Unsupervised Multimodal Representation Learning across Medical Images and Reports
Nov 21, 2018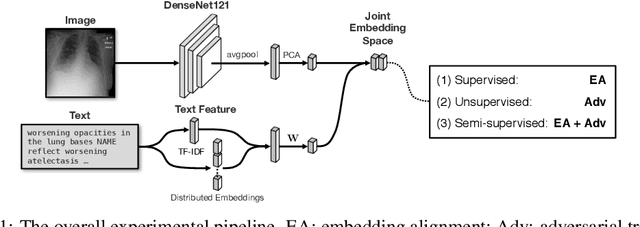
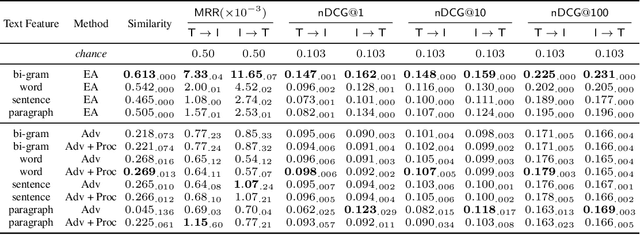
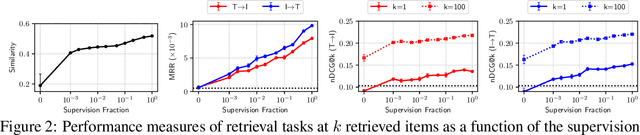
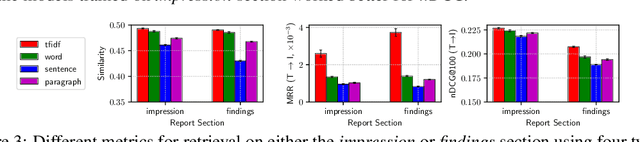
Abstract:Joint embeddings between medical imaging modalities and associated radiology reports have the potential to offer significant benefits to the clinical community, ranging from cross-domain retrieval to conditional generation of reports to the broader goals of multimodal representation learning. In this work, we establish baseline joint embedding results measured via both local and global retrieval methods on the soon to be released MIMIC-CXR dataset consisting of both chest X-ray images and the associated radiology reports. We examine both supervised and unsupervised methods on this task and show that for document retrieval tasks with the learned representations, only a limited amount of supervision is needed to yield results comparable to those of fully-supervised methods.
Modeling Mistrust in End-of-Life Care
Jun 30, 2018
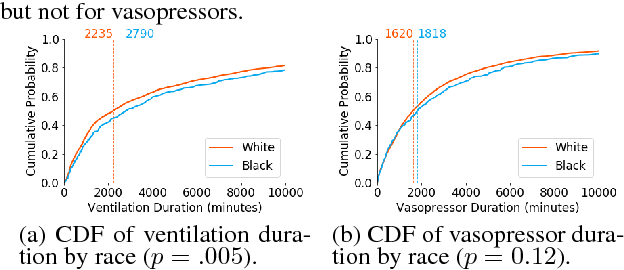
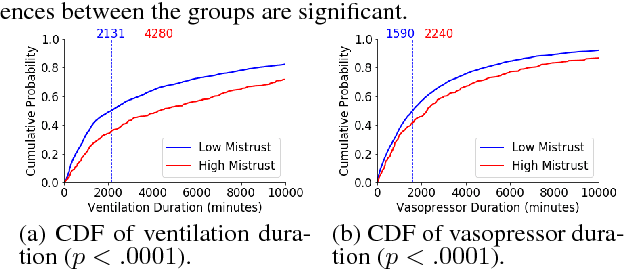
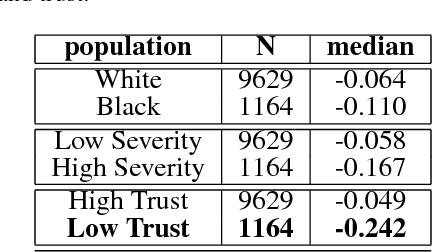
Abstract:In this work, we characterize the doctor-patient relationship using a machine learning-derived trust score. We show that this score has statistically significant racial associations, and that by modeling trust directly we find stronger disparities in care than by stratifying on race. We further demonstrate that mistrust is indicative of worse outcomes, but is only weakly associated with physiologically-created severity scores. Finally, we describe sentiment analysis experiments indicating patients with higher levels of mistrust have worse experiences and interactions with their caregivers. This work is a step towards measuring fairer machine learning in the healthcare domain.
Towards the Creation of a Large Corpus of Synthetically-Identified Clinical Notes
Mar 07, 2018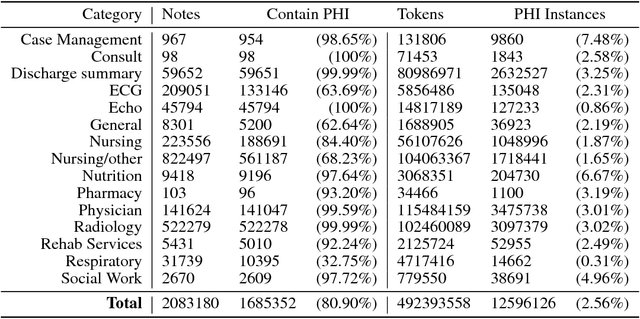
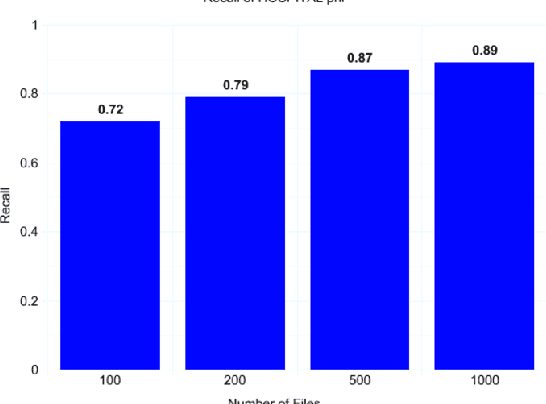
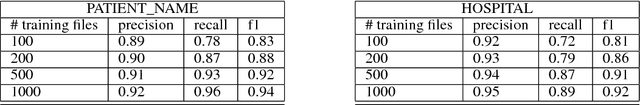
Abstract:Clinical notes often describe the most important aspects of a patient's physiology and are therefore critical to medical research. However, these notes are typically inaccessible to researchers without prior removal of sensitive protected health information (PHI), a natural language processing (NLP) task referred to as deidentification. Tools to automatically de-identify clinical notes are needed but are difficult to create without access to those very same notes containing PHI. This work presents a first step toward creating a large synthetically-identified corpus of clinical notes and corresponding PHI annotations in order to facilitate the development de-identification tools. Further, one such tool is evaluated against this corpus in order to understand the advantages and shortcomings of this approach.
CliNER 2.0: Accessible and Accurate Clinical Concept Extraction
Mar 06, 2018
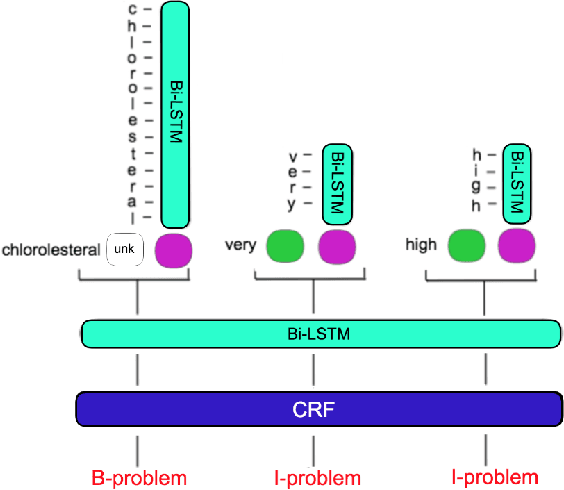

Abstract:Clinical notes often describe important aspects of a patient's stay and are therefore critical to medical research. Clinical concept extraction (CCE) of named entities - such as problems, tests, and treatments - aids in forming an understanding of notes and provides a foundation for many downstream clinical decision-making tasks. Historically, this task has been posed as a standard named entity recognition (NER) sequence tagging problem, and solved with feature-based methods using handengineered domain knowledge. Recent advances, however, have demonstrated the efficacy of LSTM-based models for NER tasks, including CCE. This work presents CliNER 2.0, a simple-to-install, open-source tool for extracting concepts from clinical text. CliNER 2.0 uses a word- and character- level LSTM model, and achieves state-of-the-art performance. For ease of use, the tool also includes pre-trained models available for public use.
AWE-CM Vectors: Augmenting Word Embeddings with a Clinical Metathesaurus
Dec 05, 2017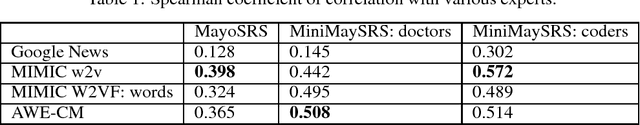
Abstract:In recent years, word embeddings have been surprisingly effective at capturing intuitive characteristics of the words they represent. These vectors achieve the best results when training corpora are extremely large, sometimes billions of words. Clinical natural language processing datasets, however, tend to be much smaller. Even the largest publicly-available dataset of medical notes is three orders of magnitude smaller than the dataset of the oft-used "Google News" word vectors. In order to make up for limited training data sizes, we encode expert domain knowledge into our embeddings. Building on a previous extension of word2vec, we show that generalizing the notion of a word's "context" to include arbitrary features creates an avenue for encoding domain knowledge into word embeddings. We show that the word vectors produced by this method outperform their text-only counterparts across the board in correlation with clinical experts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge