Wenkang Qin
EMMA: Generalizing Real-World Robot Manipulation via Generative Visual Transfer
Sep 26, 2025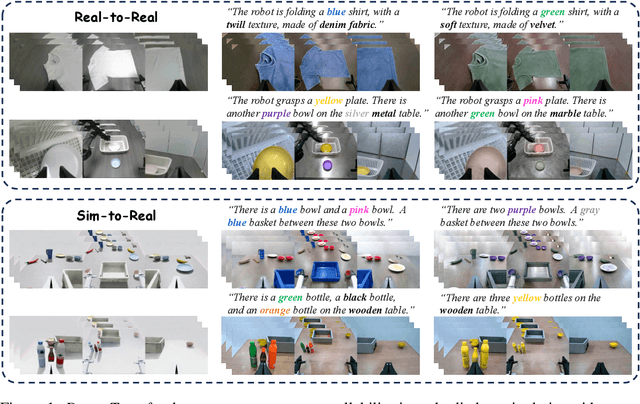
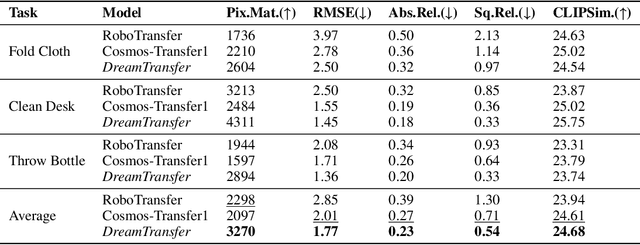
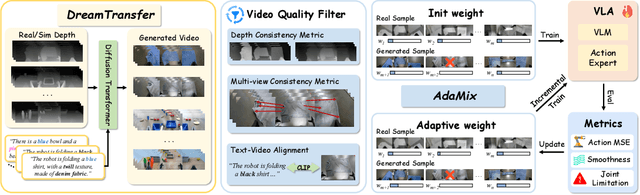
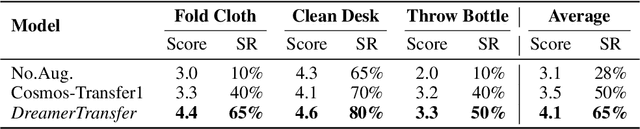
Abstract:Vision-language-action (VLA) models increasingly rely on diverse training data to achieve robust generalization. However, collecting large-scale real-world robot manipulation data across varied object appearances and environmental conditions remains prohibitively time-consuming and expensive. To overcome this bottleneck, we propose Embodied Manipulation Media Adaptation (EMMA), a VLA policy enhancement framework that integrates a generative data engine with an effective training pipeline. We introduce DreamTransfer, a diffusion Transformer-based framework for generating multi-view consistent, geometrically grounded embodied manipulation videos. DreamTransfer enables text-controlled visual editing of robot videos, transforming foreground, background, and lighting conditions without compromising 3D structure or geometrical plausibility. Furthermore, we explore hybrid training with real and generated data, and introduce AdaMix, a hard-sample-aware training strategy that dynamically reweights training batches to focus optimization on perceptually or kinematically challenging samples. Extensive experiments show that videos generated by DreamTransfer significantly outperform prior video generation methods in multi-view consistency, geometric fidelity, and text-conditioning accuracy. Crucially, VLAs trained with generated data enable robots to generalize to unseen object categories and novel visual domains using only demonstrations from a single appearance. In real-world robotic manipulation tasks with zero-shot visual domains, our approach achieves over a 200% relative performance gain compared to training on real data alone, and further improves by 13% with AdaMix, demonstrating its effectiveness in boosting policy generalization.
MimicDreamer: Aligning Human and Robot Demonstrations for Scalable VLA Training
Sep 26, 2025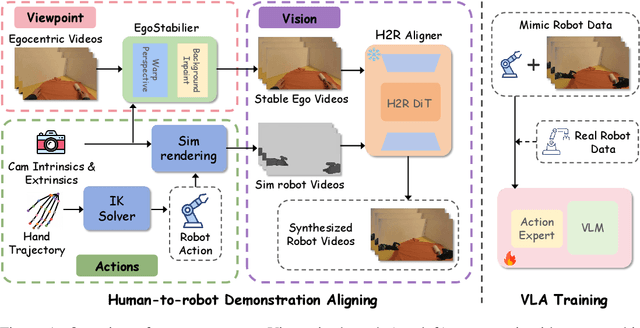
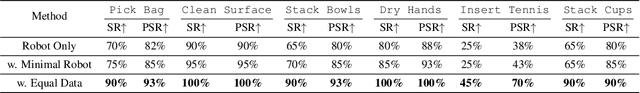
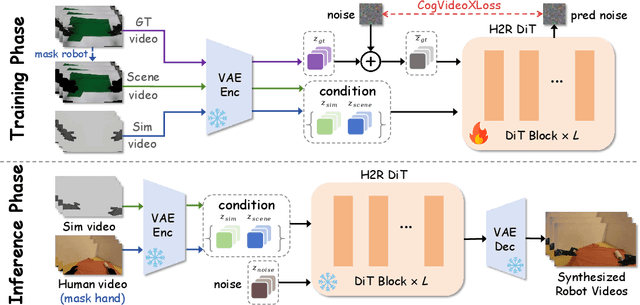
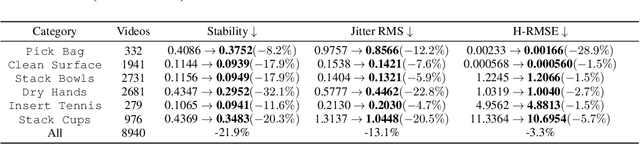
Abstract:Vision Language Action (VLA) models derive their generalization capability from diverse training data, yet collecting embodied robot interaction data remains prohibitively expensive. In contrast, human demonstration videos are far more scalable and cost-efficient to collect, and recent studies confirm their effectiveness in training VLA models. However, a significant domain gap persists between human videos and robot-executed videos, including unstable camera viewpoints, visual discrepancies between human hands and robotic arms, and differences in motion dynamics. To bridge this gap, we propose MimicDreamer, a framework that turns fast, low-cost human demonstrations into robot-usable supervision by jointly aligning vision, viewpoint, and actions to directly support policy training. For visual alignment, we propose H2R Aligner, a video diffusion model that generates high-fidelity robot demonstration videos by transferring motion from human manipulation footage. For viewpoint stabilization, EgoStabilizer is proposed, which canonicalizes egocentric videos via homography and inpaints occlusions and distortions caused by warping. For action alignment, we map human hand trajectories to the robot frame and apply a constrained inverse kinematics solver to produce feasible, low-jitter joint commands with accurate pose tracking. Empirically, VLA models trained purely on our synthesized human-to-robot videos achieve few-shot execution on real robots. Moreover, scaling training with human data significantly boosts performance compared to models trained solely on real robot data; our approach improves the average success rate by 14.7\% across six representative manipulation tasks.
ReconDreamer-RL: Enhancing Reinforcement Learning via Diffusion-based Scene Reconstruction
Aug 11, 2025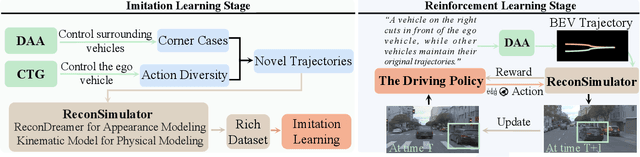


Abstract:Reinforcement learning for training end-to-end autonomous driving models in closed-loop simulations is gaining growing attention. However, most simulation environments differ significantly from real-world conditions, creating a substantial simulation-to-reality (sim2real) gap. To bridge this gap, some approaches utilize scene reconstruction techniques to create photorealistic environments as a simulator. While this improves realistic sensor simulation, these methods are inherently constrained by the distribution of the training data, making it difficult to render high-quality sensor data for novel trajectories or corner case scenarios. Therefore, we propose ReconDreamer-RL, a framework designed to integrate video diffusion priors into scene reconstruction to aid reinforcement learning, thereby enhancing end-to-end autonomous driving training. Specifically, in ReconDreamer-RL, we introduce ReconSimulator, which combines the video diffusion prior for appearance modeling and incorporates a kinematic model for physical modeling, thereby reconstructing driving scenarios from real-world data. This narrows the sim2real gap for closed-loop evaluation and reinforcement learning. To cover more corner-case scenarios, we introduce the Dynamic Adversary Agent (DAA), which adjusts the trajectories of surrounding vehicles relative to the ego vehicle, autonomously generating corner-case traffic scenarios (e.g., cut-in). Finally, the Cousin Trajectory Generator (CTG) is proposed to address the issue of training data distribution, which is often biased toward simple straight-line movements. Experiments show that ReconDreamer-RL improves end-to-end autonomous driving training, outperforming imitation learning methods with a 5x reduction in the Collision Ratio.
Humanoid Occupancy: Enabling A Generalized Multimodal Occupancy Perception System on Humanoid Robots
Jul 27, 2025Abstract:Humanoid robot technology is advancing rapidly, with manufacturers introducing diverse heterogeneous visual perception modules tailored to specific scenarios. Among various perception paradigms, occupancy-based representation has become widely recognized as particularly suitable for humanoid robots, as it provides both rich semantic and 3D geometric information essential for comprehensive environmental understanding. In this work, we present Humanoid Occupancy, a generalized multimodal occupancy perception system that integrates hardware and software components, data acquisition devices, and a dedicated annotation pipeline. Our framework employs advanced multi-modal fusion techniques to generate grid-based occupancy outputs encoding both occupancy status and semantic labels, thereby enabling holistic environmental understanding for downstream tasks such as task planning and navigation. To address the unique challenges of humanoid robots, we overcome issues such as kinematic interference and occlusion, and establish an effective sensor layout strategy. Furthermore, we have developed the first panoramic occupancy dataset specifically for humanoid robots, offering a valuable benchmark and resource for future research and development in this domain. The network architecture incorporates multi-modal feature fusion and temporal information integration to ensure robust perception. Overall, Humanoid Occupancy delivers effective environmental perception for humanoid robots and establishes a technical foundation for standardizing universal visual modules, paving the way for the widespread deployment of humanoid robots in complex real-world scenarios.
RoboTransfer: Geometry-Consistent Video Diffusion for Robotic Visual Policy Transfer
May 29, 2025Abstract:Imitation Learning has become a fundamental approach in robotic manipulation. However, collecting large-scale real-world robot demonstrations is prohibitively expensive. Simulators offer a cost-effective alternative, but the sim-to-real gap make it extremely challenging to scale. Therefore, we introduce RoboTransfer, a diffusion-based video generation framework for robotic data synthesis. Unlike previous methods, RoboTransfer integrates multi-view geometry with explicit control over scene components, such as background and object attributes. By incorporating cross-view feature interactions and global depth/normal conditions, RoboTransfer ensures geometry consistency across views. This framework allows fine-grained control, including background edits and object swaps. Experiments demonstrate that RoboTransfer is capable of generating multi-view videos with enhanced geometric consistency and visual fidelity. In addition, policies trained on the data generated by RoboTransfer achieve a 33.3% relative improvement in the success rate in the DIFF-OBJ setting and a substantial 251% relative improvement in the more challenging DIFF-ALL scenario. Explore more demos on our project page: https://horizonrobotics.github.io/robot_lab/robotransfer
WonderTurbo: Generating Interactive 3D World in 0.72 Seconds
Apr 03, 2025



Abstract:Interactive 3D generation is gaining momentum and capturing extensive attention for its potential to create immersive virtual experiences. However, a critical challenge in current 3D generation technologies lies in achieving real-time interactivity. To address this issue, we introduce WonderTurbo, the first real-time interactive 3D scene generation framework capable of generating novel perspectives of 3D scenes within 0.72 seconds. Specifically, WonderTurbo accelerates both geometric and appearance modeling in 3D scene generation. In terms of geometry, we propose StepSplat, an innovative method that constructs efficient 3D geometric representations through dynamic updates, each taking only 0.26 seconds. Additionally, we design QuickDepth, a lightweight depth completion module that provides consistent depth input for StepSplat, further enhancing geometric accuracy. For appearance modeling, we develop FastPaint, a 2-steps diffusion model tailored for instant inpainting, which focuses on maintaining spatial appearance consistency. Experimental results demonstrate that WonderTurbo achieves a remarkable 15X speedup compared to baseline methods, while preserving excellent spatial consistency and delivering high-quality output.
ReconDreamer: Crafting World Models for Driving Scene Reconstruction via Online Restoration
Nov 29, 2024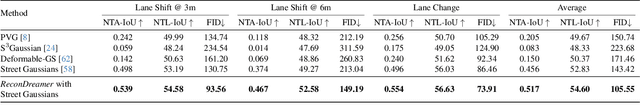



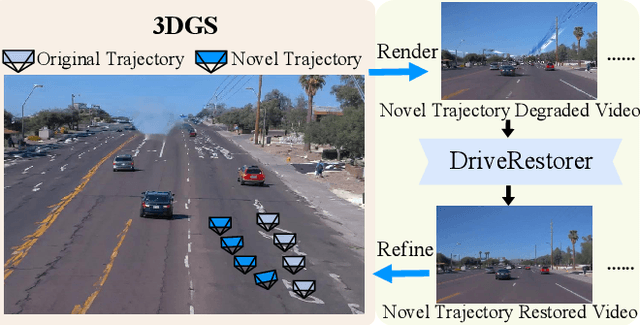
Abstract:Closed-loop simulation is crucial for end-to-end autonomous driving. Existing sensor simulation methods (e.g., NeRF and 3DGS) reconstruct driving scenes based on conditions that closely mirror training data distributions. However, these methods struggle with rendering novel trajectories, such as lane changes. Recent works have demonstrated that integrating world model knowledge alleviates these issues. Despite their efficiency, these approaches still encounter difficulties in the accurate representation of more complex maneuvers, with multi-lane shifts being a notable example. Therefore, we introduce ReconDreamer, which enhances driving scene reconstruction through incremental integration of world model knowledge. Specifically, DriveRestorer is proposed to mitigate artifacts via online restoration. This is complemented by a progressive data update strategy designed to ensure high-quality rendering for more complex maneuvers. To the best of our knowledge, ReconDreamer is the first method to effectively render in large maneuvers. Experimental results demonstrate that ReconDreamer outperforms Street Gaussians in the NTA-IoU, NTL-IoU, and FID, with relative improvements by 24.87%, 6.72%, and 29.97%. Furthermore, ReconDreamer surpasses DriveDreamer4D with PVG during large maneuver rendering, as verified by a relative improvement of 195.87% in the NTA-IoU metric and a comprehensive user study.
PathInsight: Instruction Tuning of Multimodal Datasets and Models for Intelligence Assisted Diagnosis in Histopathology
Aug 13, 2024



Abstract:Pathological diagnosis remains the definitive standard for identifying tumors. The rise of multimodal large models has simplified the process of integrating image analysis with textual descriptions. Despite this advancement, the substantial costs associated with training and deploying these complex multimodal models, together with a scarcity of high-quality training datasets, create a significant divide between cutting-edge technology and its application in the clinical setting. We had meticulously compiled a dataset of approximately 45,000 cases, covering over 6 different tasks, including the classification of organ tissues, generating pathology report descriptions, and addressing pathology-related questions and answers. We have fine-tuned multimodal large models, specifically LLaVA, Qwen-VL, InternLM, with this dataset to enhance instruction-based performance. We conducted a qualitative assessment of the capabilities of the base model and the fine-tuned model in performing image captioning and classification tasks on the specific dataset. The evaluation results demonstrate that the fine-tuned model exhibits proficiency in addressing typical pathological questions. We hope that by making both our models and datasets publicly available, they can be valuable to the medical and research communities.
SCAAT: Improving Neural Network Interpretability via Saliency Constrained Adaptive Adversarial Training
Nov 10, 2023



Abstract:Deep Neural Networks (DNNs) are expected to provide explanation for users to understand their black-box predictions. Saliency map is a common form of explanation illustrating the heatmap of feature attributions, but it suffers from noise in distinguishing important features. In this paper, we propose a model-agnostic learning method called Saliency Constrained Adaptive Adversarial Training (SCAAT) to improve the quality of such DNN interpretability. By constructing adversarial samples under the guidance of saliency map, SCAAT effectively eliminates most noise and makes saliency maps sparser and more faithful without any modification to the model architecture. We apply SCAAT to multiple DNNs and evaluate the quality of the generated saliency maps on various natural and pathological image datasets. Evaluations on different domains and metrics show that SCAAT significantly improves the interpretability of DNNs by providing more faithful saliency maps without sacrificing their predictive power.
What a Whole Slide Image Can Tell? Subtype-guided Masked Transformer for Pathological Image Captioning
Oct 31, 2023



Abstract:Pathological captioning of Whole Slide Images (WSIs), though is essential in computer-aided pathological diagnosis, has rarely been studied due to the limitations in datasets and model training efficacy. In this paper, we propose a new paradigm Subtype-guided Masked Transformer (SGMT) for pathological captioning based on Transformers, which treats a WSI as a sequence of sparse patches and generates an overall caption sentence from the sequence. An accompanying subtype prediction is introduced into SGMT to guide the training process and enhance the captioning accuracy. We also present an Asymmetric Masked Mechansim approach to tackle the large size constraint of pathological image captioning, where the numbers of sequencing patches in SGMT are sampled differently in the training and inferring phases, respectively. Experiments on the PatchGastricADC22 dataset demonstrate that our approach effectively adapts to the task with a transformer-based model and achieves superior performance than traditional RNN-based methods. Our codes are to be made available for further research and development.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge