Wenbin Hu
Variational Bayesian Flow Network for Graph Generation
Jan 30, 2026Abstract:Graph generation aims to sample discrete node and edge attributes while satisfying coupled structural constraints. Diffusion models for graphs often adopt largely factorized forward-noising, and many flow-matching methods start from factorized reference noise and coordinate-wise interpolation, so node-edge coupling is not encoded by the generative geometry and must be recovered implicitly by the core network, which can be brittle after discrete decoding. Bayesian Flow Networks (BFNs) evolve distribution parameters and naturally support discrete generation. But classical BFNs typically rely on factorized beliefs and independent channels, which limit geometric evidence fusion. We propose Variational Bayesian Flow Network (VBFN), which performs a variational lifting to a tractable joint Gaussian variational belief family governed by structured precisions. Each Bayesian update reduces to solving a symmetric positive definite linear system, enabling coupled node and edge updates within a single fusion step. We construct sample-agnostic sparse precisions from a representation-induced dependency graph, thereby avoiding label leakage while enforcing node-edge consistency. On synthetic and molecular graph datasets, VBFN improves fidelity and diversity, and surpasses baseline methods.
PCEvo: Path-Consistent Molecular Representation via Virtual Evolutionary
Jan 27, 2026Abstract:Molecular representation learning aims to learn vector embeddings that capture molecular structure and geometry, thereby enabling property prediction and downstream scientific applications. In many AI for science tasks, labeled data are expensive to obtain and therefore limited in availability. Under the few-shot setting, models trained with scarce supervision often learn brittle structure-property relationships, resulting in substantially higher prediction errors and reduced generalization to unseen molecules. To address this limitation, we propose PCEvo, a path-consistent representation method that learns from virtual paths through dynamic structural evolution. PCEvo enumerates multiple chemically feasible edit paths between retrieved similar molecular pairs under topological dependency constraints. It transforms the labels of the two molecules into stepwise supervision along each virtual evolutionary path. It introduces a path-consistency objective that enforces prediction invariance across alternative paths connecting the same two molecules. Comprehensive experiments on the QM9 and MoleculeNet datasets demonstrate that PCEvo substantially improves the few-shot generalization performance of baseline methods. The code is available at https://anonymous.4open.science/r/PCEvo-4BF2.
Safety Compliance: Rethinking LLM Safety Reasoning through the Lens of Compliance
Sep 26, 2025Abstract:The proliferation of Large Language Models (LLMs) has demonstrated remarkable capabilities, elevating the critical importance of LLM safety. However, existing safety methods rely on ad-hoc taxonomy and lack a rigorous, systematic protection, failing to ensure safety for the nuanced and complex behaviors of modern LLM systems. To address this problem, we solve LLM safety from legal compliance perspectives, named safety compliance. In this work, we posit relevant established legal frameworks as safety standards for defining and measuring safety compliance, including the EU AI Act and GDPR, which serve as core legal frameworks for AI safety and data security in Europe. To bridge the gap between LLM safety and legal compliance, we first develop a new benchmark for safety compliance by generating realistic LLM safety scenarios seeded with legal statutes. Subsequently, we align Qwen3-8B using Group Policy Optimization (GRPO) to construct a safety reasoner, Compliance Reasoner, which effectively aligns LLMs with legal standards to mitigate safety risks. Our comprehensive experiments demonstrate that the Compliance Reasoner achieves superior performance on the new benchmark, with average improvements of +10.45% for the EU AI Act and +11.85% for GDPR.
Zero-Shot Learning with Subsequence Reordering Pretraining for Compound-Protein Interaction
Jul 28, 2025Abstract:Given the vastness of chemical space and the ongoing emergence of previously uncharacterized proteins, zero-shot compound-protein interaction (CPI) prediction better reflects the practical challenges and requirements of real-world drug development. Although existing methods perform adequately during certain CPI tasks, they still face the following challenges: (1) Representation learning from local or complete protein sequences often overlooks the complex interdependencies between subsequences, which are essential for predicting spatial structures and binding properties. (2) Dependence on large-scale or scarce multimodal protein datasets demands significant training data and computational resources, limiting scalability and efficiency. To address these challenges, we propose a novel approach that pretrains protein representations for CPI prediction tasks using subsequence reordering, explicitly capturing the dependencies between protein subsequences. Furthermore, we apply length-variable protein augmentation to ensure excellent pretraining performance on small training datasets. To evaluate the model's effectiveness and zero-shot learning ability, we combine it with various baseline methods. The results demonstrate that our approach can improve the baseline model's performance on the CPI task, especially in the challenging zero-shot scenario. Compared to existing pre-training models, our model demonstrates superior performance, particularly in data-scarce scenarios where training samples are limited. Our implementation is available at https://github.com/Hoch-Zhang/PSRP-CPI.
Antibody Design and Optimization with Multi-scale Equivariant Graph Diffusion Models for Accurate Complex Antigen Binding
Jun 26, 2025Abstract:Antibody design remains a critical challenge in therapeutic and diagnostic development, particularly for complex antigens with diverse binding interfaces. Current computational methods face two main limitations: (1) capturing geometric features while preserving symmetries, and (2) generalizing novel antigen interfaces. Despite recent advancements, these methods often fail to accurately capture molecular interactions and maintain structural integrity. To address these challenges, we propose \textbf{AbMEGD}, an end-to-end framework integrating \textbf{M}ulti-scale \textbf{E}quivariant \textbf{G}raph \textbf{D}iffusion for antibody sequence and structure co-design. Leveraging advanced geometric deep learning, AbMEGD combines atomic-level geometric features with residue-level embeddings, capturing local atomic details and global sequence-structure interactions. Its E(3)-equivariant diffusion method ensures geometric precision, computational efficiency, and robust generalizability for complex antigens. Furthermore, experiments using the SAbDab database demonstrate a 10.13\% increase in amino acid recovery, 3.32\% rise in improvement percentage, and a 0.062~\AA\ reduction in root mean square deviation within the critical CDR-H3 region compared to DiffAb, a leading antibody design model. These results highlight AbMEGD's ability to balance structural integrity with improved functionality, establishing a new benchmark for sequence-structure co-design and affinity optimization. The code is available at: https://github.com/Patrick221215/AbMEGD.
MCIP: Protecting MCP Safety via Model Contextual Integrity Protocol
May 21, 2025Abstract:As Model Context Protocol (MCP) introduces an easy-to-use ecosystem for users and developers, it also brings underexplored safety risks. Its decentralized architecture, which separates clients and servers, poses unique challenges for systematic safety analysis. This paper proposes a novel framework to enhance MCP safety. Guided by the MAESTRO framework, we first analyze the missing safety mechanisms in MCP, and based on this analysis, we propose the Model Contextual Integrity Protocol (MCIP), a refined version of MCP that addresses these gaps. Next, we develop a fine-grained taxonomy that captures a diverse range of unsafe behaviors observed in MCP scenarios. Building on this taxonomy, we develop benchmark and training data that support the evaluation and improvement of LLMs' capabilities in identifying safety risks within MCP interactions. Leveraging the proposed benchmark and training data, we conduct extensive experiments on state-of-the-art LLMs. The results highlight LLMs' vulnerabilities in MCP interactions and demonstrate that our approach substantially improves their safety performance.
DrugPilot: LLM-based Parameterized Reasoning Agent for Drug Discovery
May 20, 2025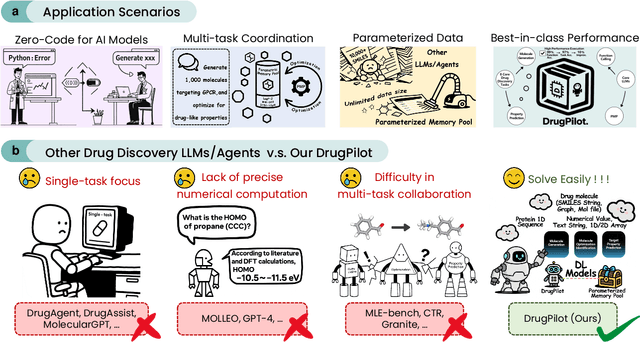
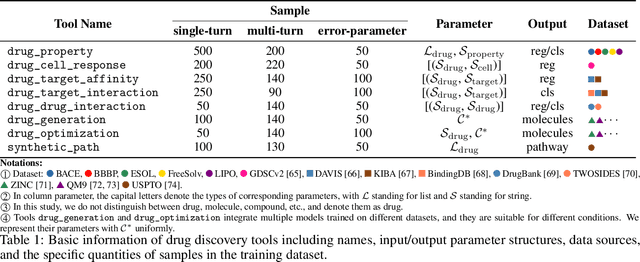
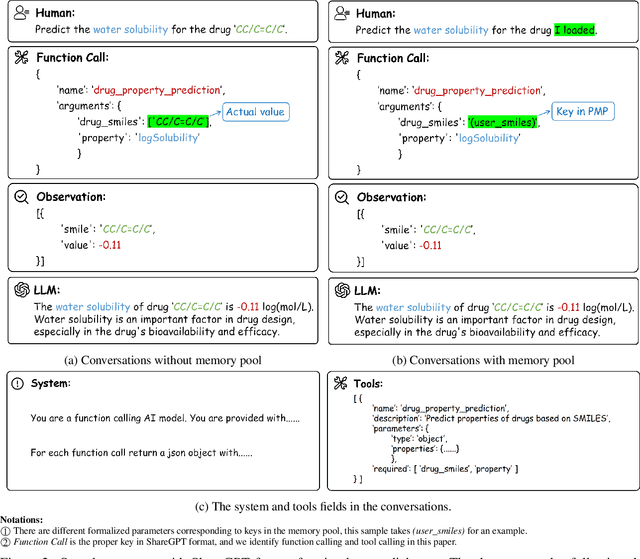
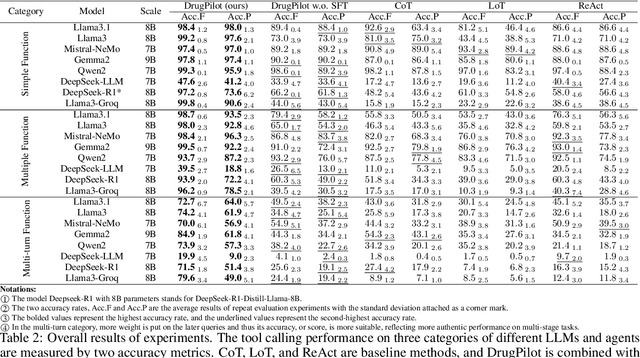
Abstract:In the field of AI4Science, large-scale language models (LLMs) show great potential to parse complex scientific semantics, integrate cross-disciplinary knowledge, and assist critical task research. However, in the field of drug discovery, despite the optimization through professional data pre-training, context window expansion, and internet search, the existing LLMs are still facing challenges such as massive multi-modal and heterogeneous data processing, domain knowledge dynamic updating delay, and insufficient confidence in predicting the results of complex computational tasks. To address these challenges, we propose the DrugPilot, an LLM-based agent with parameterized reasoning for drug discovery. DrugPilot addresses key limitations of traditional end-to-end LLM prediction approaches through its parametric inference architecture. This agent system supports major phases of the drug discovery pipeline, facilitating automated planning and execution of multi-stage research tasks. To address the critical challenge of multi-modal drug data analysis (incorporating both public datasets and user-submitted data), we developed an interactive parameterized memory pool. This innovative component standardizes real-world drug data into parametric representations, simultaneously enabling efficient knowledge retrieval in multi-turn dialogue while mitigating the information loss inherent in text-based data transmission. Additionally, we created a drug instruct dataset across 8 essential drug discovery tasks for model fine-tuning and evaluation. Based on the Berkeley function calling evaluation framework, DrugPilot demonstrated the most advanced tool calling capabilities on our drug discovery tool instruction dataset, outperforming existing agents (e.g., ReAct, LoT). Specifically, it achieves task completion rates of 98.0%, 93.5%, and 64.0% on simple, multiple, and multi-turn tasks, respectively.
Context Reasoner: Incentivizing Reasoning Capability for Contextualized Privacy and Safety Compliance via Reinforcement Learning
May 20, 2025Abstract:While Large Language Models (LLMs) exhibit remarkable capabilities, they also introduce significant safety and privacy risks. Current mitigation strategies often fail to preserve contextual reasoning capabilities in risky scenarios. Instead, they rely heavily on sensitive pattern matching to protect LLMs, which limits the scope. Furthermore, they overlook established safety and privacy standards, leading to systemic risks for legal compliance. To address these gaps, we formulate safety and privacy issues into contextualized compliance problems following the Contextual Integrity (CI) theory. Under the CI framework, we align our model with three critical regulatory standards: GDPR, EU AI Act, and HIPAA. Specifically, we employ reinforcement learning (RL) with a rule-based reward to incentivize contextual reasoning capabilities while enhancing compliance with safety and privacy norms. Through extensive experiments, we demonstrate that our method not only significantly enhances legal compliance (achieving a +17.64% accuracy improvement in safety/privacy benchmarks) but also further improves general reasoning capability. For OpenThinker-7B, a strong reasoning model that significantly outperforms its base model Qwen2.5-7B-Instruct across diverse subjects, our method enhances its general reasoning capabilities, with +2.05% and +8.98% accuracy improvement on the MMLU and LegalBench benchmark, respectively.
Benchmarking Reasoning Robustness in Large Language Models
Mar 06, 2025Abstract:Despite the recent success of large language models (LLMs) in reasoning such as DeepSeek, we for the first time identify a key dilemma in reasoning robustness and generalization: significant performance degradation on novel or incomplete data, suggesting a reliance on memorized patterns rather than systematic reasoning. Our closer examination reveals four key unique limitations underlying this issue:(1) Positional bias--models favor earlier queries in multi-query inputs but answering the wrong one in the latter (e.g., GPT-4o's accuracy drops from 75.8 percent to 72.8 percent); (2) Instruction sensitivity--performance declines by 5.0 to 7.5 percent in the Qwen2.5 Series and by 5.0 percent in DeepSeek-V3 with auxiliary guidance; (3) Numerical fragility--value substitution sharply reduces accuracy (e.g., GPT-4o drops from 97.5 percent to 82.5 percent, GPT-o1-mini drops from 97.5 percent to 92.5 percent); and (4) Memory dependence--models resort to guesswork when missing critical data. These findings further highlight the reliance on heuristic recall over rigorous logical inference, demonstrating challenges in reasoning robustness. To comprehensively investigate these robustness challenges, this paper introduces a novel benchmark, termed as Math-RoB, that exploits hallucinations triggered by missing information to expose reasoning gaps. This is achieved by an instruction-based approach to generate diverse datasets that closely resemble training distributions, facilitating a holistic robustness assessment and advancing the development of more robust reasoning frameworks. Bad character(s) in field Abstract.
Graph-Augmented Reasoning: Evolving Step-by-Step Knowledge Graph Retrieval for LLM Reasoning
Mar 03, 2025



Abstract:Recent large language model (LLM) reasoning, despite its success, suffers from limited domain knowledge, susceptibility to hallucinations, and constrained reasoning depth, particularly in small-scale models deployed in resource-constrained environments. This paper presents the first investigation into integrating step-wise knowledge graph retrieval with step-wise reasoning to address these challenges, introducing a novel paradigm termed as graph-augmented reasoning. Our goal is to enable frozen, small-scale LLMs to retrieve and process relevant mathematical knowledge in a step-wise manner, enhancing their problem-solving abilities without additional training. To this end, we propose KG-RAR, a framework centered on process-oriented knowledge graph construction, a hierarchical retrieval strategy, and a universal post-retrieval processing and reward model (PRP-RM) that refines retrieved information and evaluates each reasoning step. Experiments on the Math500 and GSM8K benchmarks across six models demonstrate that KG-RAR yields encouraging results, achieving a 20.73\% relative improvement with Llama-3B on Math500.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge