Xiuwen Gong
Variational Bayesian Flow Network for Graph Generation
Jan 30, 2026Abstract:Graph generation aims to sample discrete node and edge attributes while satisfying coupled structural constraints. Diffusion models for graphs often adopt largely factorized forward-noising, and many flow-matching methods start from factorized reference noise and coordinate-wise interpolation, so node-edge coupling is not encoded by the generative geometry and must be recovered implicitly by the core network, which can be brittle after discrete decoding. Bayesian Flow Networks (BFNs) evolve distribution parameters and naturally support discrete generation. But classical BFNs typically rely on factorized beliefs and independent channels, which limit geometric evidence fusion. We propose Variational Bayesian Flow Network (VBFN), which performs a variational lifting to a tractable joint Gaussian variational belief family governed by structured precisions. Each Bayesian update reduces to solving a symmetric positive definite linear system, enabling coupled node and edge updates within a single fusion step. We construct sample-agnostic sparse precisions from a representation-induced dependency graph, thereby avoiding label leakage while enforcing node-edge consistency. On synthetic and molecular graph datasets, VBFN improves fidelity and diversity, and surpasses baseline methods.
Knowledge-aware contrastive heterogeneous molecular graph learning
Feb 17, 2025Abstract:Molecular representation learning is pivotal in predicting molecular properties and advancing drug design. Traditional methodologies, which predominantly rely on homogeneous graph encoding, are limited by their inability to integrate external knowledge and represent molecular structures across different levels of granularity. To address these limitations, we propose a paradigm shift by encoding molecular graphs into heterogeneous structures, introducing a novel framework: Knowledge-aware Contrastive Heterogeneous Molecular Graph Learning (KCHML). This approach leverages contrastive learning to enrich molecular representations with embedded external knowledge. KCHML conceptualizes molecules through three distinct graph views-molecular, elemental, and pharmacological-enhanced by heterogeneous molecular graphs and a dual message-passing mechanism. This design offers a comprehensive representation for property prediction, as well as for downstream tasks such as drug-drug interaction (DDI) prediction. Extensive benchmarking demonstrates KCHML's superiority over state-of-the-art molecular property prediction models, underscoring its ability to capture intricate molecular features.
A Cross-Field Fusion Strategy for Drug-Target Interaction Prediction
May 23, 2024



Abstract:Drug-target interaction (DTI) prediction is a critical component of the drug discovery process. In the drug development engineering field, predicting novel drug-target interactions is extremely crucial.However, although existing methods have achieved high accuracy levels in predicting known drugs and drug targets, they fail to utilize global protein information during DTI prediction. This leads to an inability to effectively predict interaction the interactions between novel drugs and their targets. As a result, the cross-field information fusion strategy is employed to acquire local and global protein information. Thus, we propose the siamese drug-target interaction SiamDTI prediction method, which utilizes a double channel network structure for cross-field supervised learning.Experimental results on three benchmark datasets demonstrate that SiamDTI achieves higher accuracy levels than other state-of-the-art (SOTA) methods on novel drugs and targets.Additionally, SiamDTI's performance with known drugs and targets is comparable to that of SOTA approachs. The code is available at https://anonymous.4open.science/r/DDDTI-434D.
Regressor-free Molecule Generation to Support Drug Response Prediction
May 23, 2024Abstract:Drug response prediction (DRP) is a crucial phase in drug discovery, and the most important metric for its evaluation is the IC50 score. DRP results are heavily dependent on the quality of the generated molecules. Existing molecule generation methods typically employ classifier-based guidance, enabling sampling within the IC50 classification range. However, these methods fail to ensure the sampling space range's effectiveness, generating numerous ineffective molecules. Through experimental and theoretical study, we hypothesize that conditional generation based on the target IC50 score can obtain a more effective sampling space. As a result, we introduce regressor-free guidance molecule generation to ensure sampling within a more effective space and support DRP. Regressor-free guidance combines a diffusion model's score estimation with a regression controller model's gradient based on number labels. To effectively map regression labels between drugs and cell lines, we design a common-sense numerical knowledge graph that constrains the order of text representations. Experimental results on the real-world dataset for the DRP task demonstrate our method's effectiveness in drug discovery. The code is available at:https://anonymous.4open.science/r/RMCD-DBD1.
Fast Multi-label Learning
Aug 31, 2021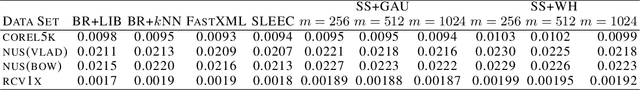
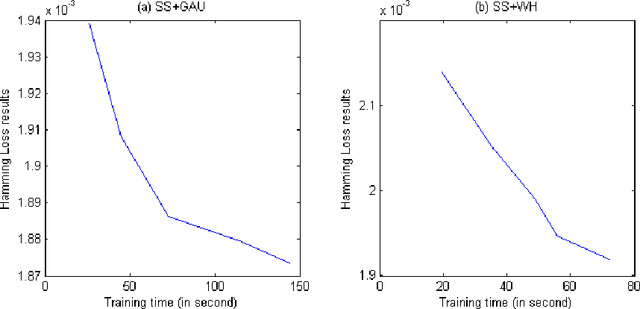
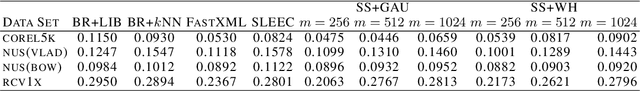
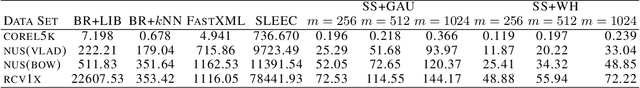
Abstract:Embedding approaches have become one of the most pervasive techniques for multi-label classification. However, the training process of embedding methods usually involves a complex quadratic or semidefinite programming problem, or the model may even involve an NP-hard problem. Thus, such methods are prohibitive on large-scale applications. More importantly, much of the literature has already shown that the binary relevance (BR) method is usually good enough for some applications. Unfortunately, BR runs slowly due to its linear dependence on the size of the input data. The goal of this paper is to provide a simple method, yet with provable guarantees, which can achieve competitive performance without a complex training process. To achieve our goal, we provide a simple stochastic sketch strategy for multi-label classification and present theoretical results from both algorithmic and statistical learning perspectives. Our comprehensive empirical studies corroborate our theoretical findings and demonstrate the superiority of the proposed methods.
Online Metric Learning for Multi-Label Classification
Jun 12, 2020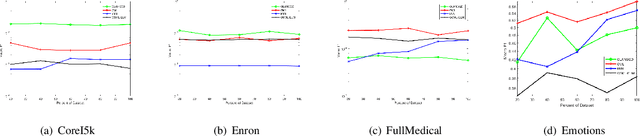
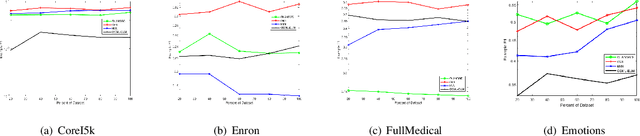
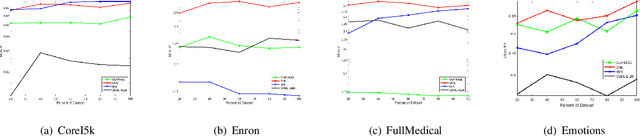

Abstract:Existing research into online multi-label classification, such as online sequential multi-label extreme learning machine (OSML-ELM) and stochastic gradient descent (SGD), has achieved promising performance. However, these works do not take label dependencies into consideration and lack a theoretical analysis of loss functions. Accordingly, we propose a novel online metric learning paradigm for multi-label classification to fill the current research gap. Generally, we first propose a new metric for multi-label classification which is based on $k$-Nearest Neighbour ($k$NN) and combined with large margin principle. Then, we adapt it to the online settting to derive our model which deals with massive volume ofstreaming data at a higher speed online. Specifically, in order to learn the new $k$NN-based metric, we first project instances in the training dataset into the label space, which make it possible for the comparisons of instances and labels in the same dimension. After that, we project both of them into a new lower dimension space simultaneously, which enables us to extract the structure of dependencies between instances and labels. Finally, we leverage the large margin and $k$NN principle to learn the metric with an efficient optimization algorithm. Moreover, we provide theoretical analysis on the upper bound of the cumulative loss for our method. Comprehensive experiments on a number of benchmark multi-label datasets validate our theoretical approach and illustrate that our proposed online metric learning (OML) algorithm outperforms state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge