Weixin Si
TempDiffReg: Temporal Diffusion Model for Non-Rigid 2D-3D Vascular Registration
Jan 26, 2026Abstract:Transarterial chemoembolization (TACE) is a preferred treatment option for hepatocellular carcinoma and other liver malignancies, yet it remains a highly challenging procedure due to complex intra-operative vascular navigation and anatomical variability. Accurate and robust 2D-3D vessel registration is essential to guide microcatheter and instruments during TACE, enabling precise localization of vascular structures and optimal therapeutic targeting. To tackle this issue, we develop a coarse-to-fine registration strategy. First, we introduce a global alignment module, structure-aware perspective n-point (SA-PnP), to establish correspondence between 2D and 3D vessel structures. Second, we propose TempDiffReg, a temporal diffusion model that performs vessel deformation iteratively by leveraging temporal context to capture complex anatomical variations and local structural changes. We collected data from 23 patients and constructed 626 paired multi-frame samples for comprehensive evaluation. Experimental results demonstrate that the proposed method consistently outperforms state-of-the-art (SOTA) methods in both accuracy and anatomical plausibility. Specifically, our method achieves a mean squared error (MSE) of 0.63 mm and a mean absolute error (MAE) of 0.51 mm in registration accuracy, representing 66.7\% lower MSE and 17.7\% lower MAE compared to the most competitive existing approaches. It has the potential to assist less-experienced clinicians in safely and efficiently performing complex TACE procedures, ultimately enhancing both surgical outcomes and patient care. Code and data are available at: \textcolor{blue}{https://github.com/LZH970328/TempDiffReg.git}
PAS-Mamba: Phase-Amplitude-Spatial State Space Model for MRI Reconstruction
Jan 20, 2026Abstract:Joint feature modeling in both the spatial and frequency domains has become a mainstream approach in MRI reconstruction. However, existing methods generally treat the frequency domain as a whole, neglecting the differences in the information carried by its internal components. According to Fourier transform theory, phase and amplitude represent different types of information in the image. Our spectrum swapping experiments show that magnitude mainly reflects pixel-level intensity, while phase predominantly governs image structure. To prevent interference between phase and magnitude feature learning caused by unified frequency-domain modeling, we propose the Phase-Amplitude-Spatial State Space Model (PAS-Mamba) for MRI Reconstruction, a framework that decouples phase and magnitude modeling in the frequency domain and combines it with image-domain features for better reconstruction. In the image domain, LocalMamba preserves spatial locality to sharpen fine anatomical details. In frequency domain, we disentangle amplitude and phase into two specialized branches to avoid representational coupling. To respect the concentric geometry of frequency information, we propose Circular Frequency Domain Scanning (CFDS) to serialize features from low to high frequencies. Finally, a Dual-Domain Complementary Fusion Module (DDCFM) adaptively fuses amplitude phase representations and enables bidirectional exchange between frequency and image domains, delivering superior reconstruction. Extensive experiments on the IXI and fastMRI knee datasets show that PAS-Mamba consistently outperforms state of the art reconstruction methods.
Surgical Scene Segmentation using a Spike-Driven Video Transformer with Real-Time Potential
Dec 24, 2025Abstract:Modern surgical systems increasingly rely on intelligent scene understanding to provide timely situational awareness for enhanced intra-operative safety. Within this pipeline, surgical scene segmentation plays a central role in accurately perceiving operative events. Although recent deep learning models, particularly large-scale foundation models, achieve remarkable segmentation accuracy, their substantial computational demands and power consumption hinder real-time deployment in resource-constrained surgical environments. To address this limitation, we explore the emerging SNN as a promising paradigm for highly efficient surgical intelligence. However, their performance is still constrained by the scarcity of labeled surgical data and the inherently sparse nature of surgical video representations. To this end, we propose \textit{SpikeSurgSeg}, the first spike-driven video Transformer framework tailored for surgical scene segmentation with real-time potential on non-GPU platforms. To address the limited availability of surgical annotations, we introduce a surgical-scene masked autoencoding pretraining strategy for SNNs that enables robust spatiotemporal representation learning via layer-wise tube masking. Building on this pretrained backbone, we further adopt a lightweight spike-driven segmentation head that produces temporally consistent predictions while preserving the low-latency characteristics of SNNs. Extensive experiments on EndoVis18 and our in-house SurgBleed dataset demonstrate that SpikeSurgSeg achieves mIoU comparable to SOTA ANN-based models while reducing inference latency by at least $8\times$. Notably, it delivers over $20\times$ acceleration relative to most foundation-model baselines, underscoring its potential for time-critical surgical scene segmentation.
Versatile and Efficient Medical Image Super-Resolution Via Frequency-Gated Mamba
Oct 31, 2025Abstract:Medical image super-resolution (SR) is essential for enhancing diagnostic accuracy while reducing acquisition cost and scanning time. However, modeling both long-range anatomical structures and fine-grained frequency details with low computational overhead remains challenging. We propose FGMamba, a novel frequency-aware gated state-space model that unifies global dependency modeling and fine-detail enhancement into a lightweight architecture. Our method introduces two key innovations: a Gated Attention-enhanced State-Space Module (GASM) that integrates efficient state-space modeling with dual-branch spatial and channel attention, and a Pyramid Frequency Fusion Module (PFFM) that captures high-frequency details across multiple resolutions via FFT-guided fusion. Extensive evaluations across five medical imaging modalities (Ultrasound, OCT, MRI, CT, and Endoscopic) demonstrate that FGMamba achieves superior PSNR/SSIM while maintaining a compact parameter footprint ($<$0.75M), outperforming CNN-based and Transformer-based SOTAs. Our results validate the effectiveness of frequency-aware state-space modeling for scalable and accurate medical image enhancement.
TFKAN: Time-Frequency KAN for Long-Term Time Series Forecasting
Jun 15, 2025Abstract:Kolmogorov-Arnold Networks (KANs) are highly effective in long-term time series forecasting due to their ability to efficiently represent nonlinear relationships and exhibit local plasticity. However, prior research on KANs has predominantly focused on the time domain, neglecting the potential of the frequency domain. The frequency domain of time series data reveals recurring patterns and periodic behaviors, which complement the temporal information captured in the time domain. To address this gap, we explore the application of KANs in the frequency domain for long-term time series forecasting. By leveraging KANs' adaptive activation functions and their comprehensive representation of signals in the frequency domain, we can more effectively learn global dependencies and periodic patterns. To integrate information from both time and frequency domains, we propose the $\textbf{T}$ime-$\textbf{F}$requency KAN (TFKAN). TFKAN employs a dual-branch architecture that independently processes features from each domain, ensuring that the distinct characteristics of each domain are fully utilized without interference. Additionally, to account for the heterogeneity between domains, we introduce a dimension-adjustment strategy that selectively upscales only in the frequency domain, enhancing efficiency while capturing richer frequency information. Experimental results demonstrate that TFKAN consistently outperforms state-of-the-art (SOTA) methods across multiple datasets. The code is available at https://github.com/LcWave/TFKAN.
A Comprehensive Survey on Magnetic Resonance Image Reconstruction
Mar 10, 2025



Abstract:Magnetic resonance imaging (MRI) reconstruction is a fundamental task aimed at recovering high-quality images from undersampled or low-quality MRI data. This process enhances diagnostic accuracy and optimizes clinical applications. In recent years, deep learning-based MRI reconstruction has made significant progress. Advancements include single-modality feature extraction using different network architectures, the integration of multimodal information, and the adoption of unsupervised or semi-supervised learning strategies. However, despite extensive research, MRI reconstruction remains a challenging problem that has yet to be fully resolved. This survey provides a systematic review of MRI reconstruction methods, covering key aspects such as data acquisition and preprocessing, publicly available datasets, single and multi-modal reconstruction models, training strategies, and evaluation metrics based on image reconstruction and downstream tasks. Additionally, we analyze the major challenges in this field and explore potential future directions.
Surgical Workflow Recognition and Blocking Effectiveness Detection in Laparoscopic Liver Resections with Pringle Maneuver
Aug 21, 2024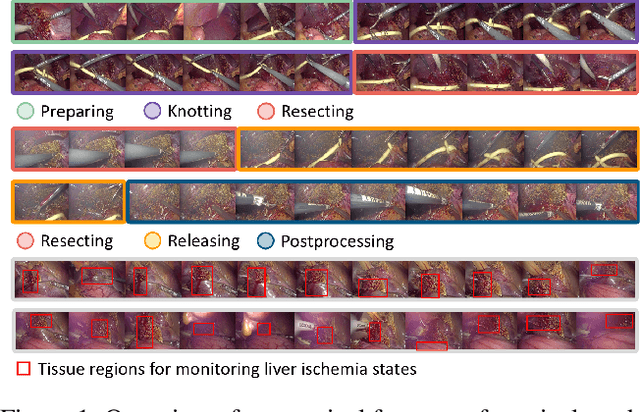

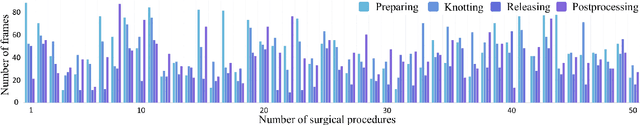

Abstract:Pringle maneuver (PM) in laparoscopic liver resection aims to reduce blood loss and provide a clear surgical view by intermittently blocking blood inflow of the liver, whereas prolonged PM may cause ischemic injury. To comprehensively monitor this surgical procedure and provide timely warnings of ineffective and prolonged blocking, we suggest two complementary AI-assisted surgical monitoring tasks: workflow recognition and blocking effectiveness detection in liver resections. The former presents challenges in real-time capturing of short-term PM, while the latter involves the intraoperative discrimination of long-term liver ischemia states. To address these challenges, we meticulously collect a novel dataset, called PmLR50, consisting of 25,037 video frames covering various surgical phases from 50 laparoscopic liver resection procedures. Additionally, we develop an online baseline for PmLR50, termed PmNet. This model embraces Masked Temporal Encoding (MTE) and Compressed Sequence Modeling (CSM) for efficient short-term and long-term temporal information modeling, and embeds Contrastive Prototype Separation (CPS) to enhance action discrimination between similar intraoperative operations. Experimental results demonstrate that PmNet outperforms existing state-of-the-art surgical workflow recognition methods on the PmLR50 benchmark. Our research offers potential clinical applications for the laparoscopic liver surgery community. Source code and data will be publicly available.
Depth-Driven Geometric Prompt Learning for Laparoscopic Liver Landmark Detection
Jun 27, 2024



Abstract:Laparoscopic liver surgery poses a complex intraoperative dynamic environment for surgeons, where remains a significant challenge to distinguish critical or even hidden structures inside the liver. Liver anatomical landmarks, e.g., ridge and ligament, serve as important markers for 2D-3D alignment, which can significantly enhance the spatial perception of surgeons for precise surgery. To facilitate the detection of laparoscopic liver landmarks, we collect a novel dataset called L3D, which comprises 1,152 frames with elaborated landmark annotations from surgical videos of 39 patients across two medical sites. For benchmarking purposes, 12 mainstream detection methods are selected and comprehensively evaluated on L3D. Further, we propose a depth-driven geometric prompt learning network, namely D2GPLand. Specifically, we design a Depth-aware Prompt Embedding (DPE) module that is guided by self-supervised prompts and generates semantically relevant geometric information with the benefit of global depth cues extracted from SAM-based features. Additionally, a Semantic-specific Geometric Augmentation (SGA) scheme is introduced to efficiently merge RGB-D spatial and geometric information through reverse anatomic perception. The experimental results indicate that D2GPLand obtains state-of-the-art performance on L3D, with 63.52% DICE and 48.68% IoU scores. Together with 2D-3D fusion technology, our method can directly provide the surgeon with intuitive guidance information in laparoscopic scenarios.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020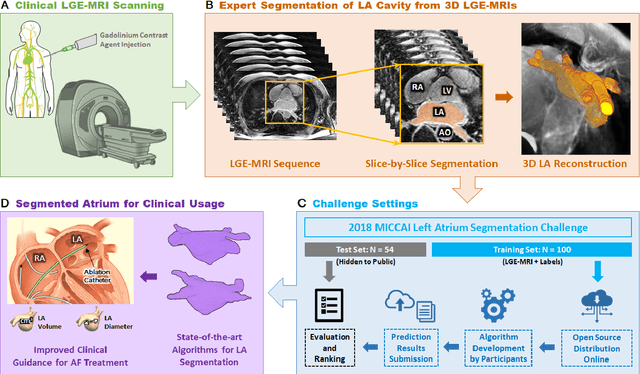
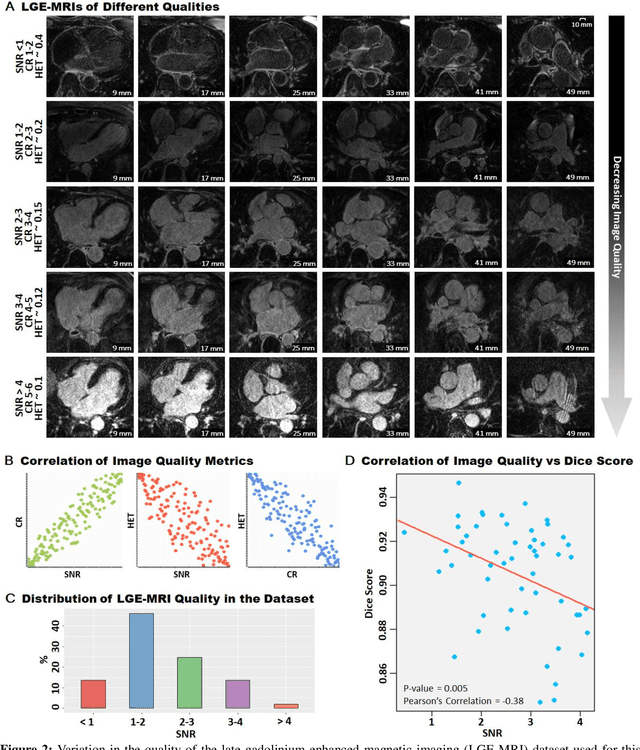
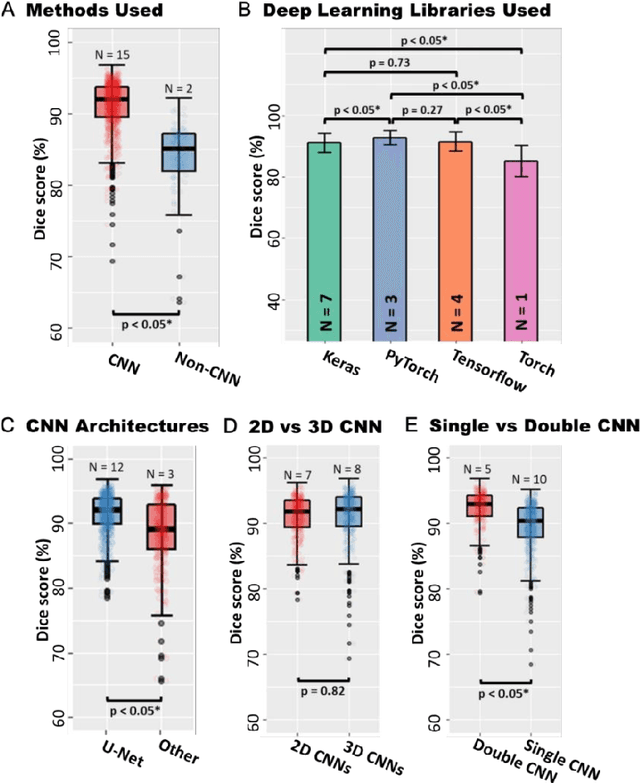
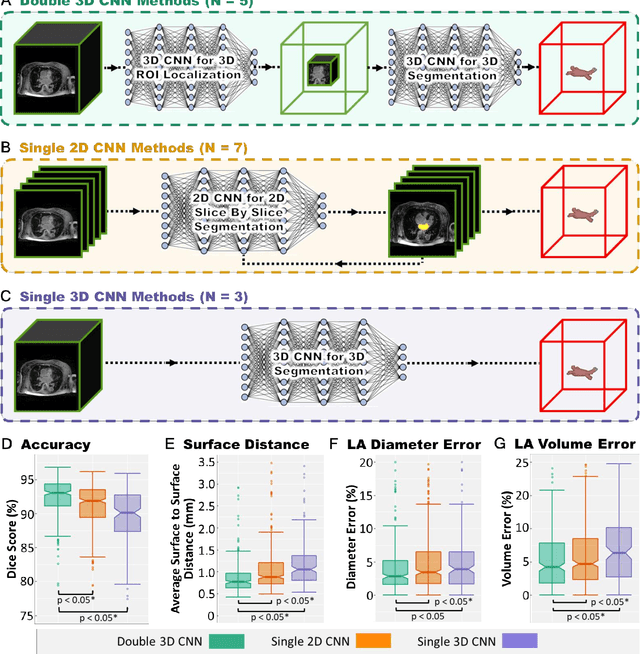
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
Evaluation of Algorithms for Multi-Modality Whole Heart Segmentation: An Open-Access Grand Challenge
Feb 21, 2019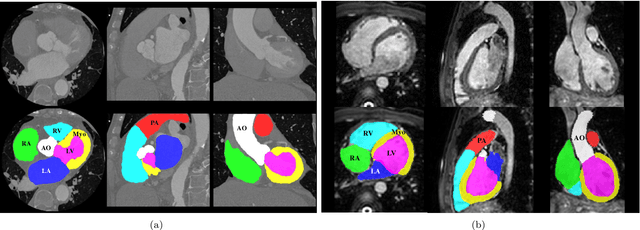
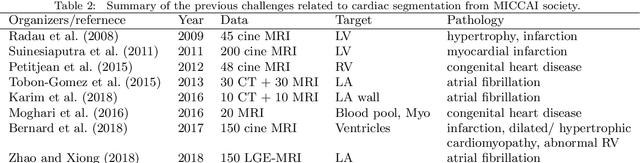
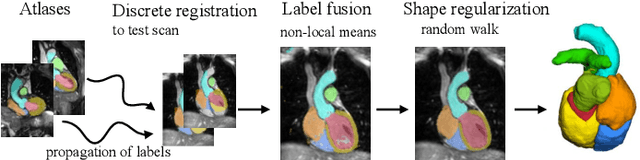
Abstract:Knowledge of whole heart anatomy is a prerequisite for many clinical applications. Whole heart segmentation (WHS), which delineates substructures of the heart, can be very valuable for modeling and analysis of the anatomy and functions of the heart. However, automating this segmentation can be arduous due to the large variation of the heart shape, and different image qualities of the clinical data. To achieve this goal, a set of training data is generally needed for constructing priors or for training. In addition, it is difficult to perform comparisons between different methods, largely due to differences in the datasets and evaluation metrics used. This manuscript presents the methodologies and evaluation results for the WHS algorithms selected from the submissions to the Multi-Modality Whole Heart Segmentation (MM-WHS) challenge, in conjunction with MICCAI 2017. The challenge provides 120 three-dimensional cardiac images covering the whole heart, including 60 CT and 60 MRI volumes, all acquired in clinical environments with manual delineation. Ten algorithms for CT data and eleven algorithms for MRI data, submitted from twelve groups, have been evaluated. The results show that many of the deep learning (DL) based methods achieved high accuracy, even though the number of training datasets was limited. A number of them also reported poor results in the blinded evaluation, probably due to overfitting in their training. The conventional algorithms, mainly based on multi-atlas segmentation, demonstrated robust and stable performance, even though the accuracy is not as good as the best DL method in CT segmentation. The challenge, including the provision of the annotated training data and the blinded evaluation for submitted algorithms on the test data, continues as an ongoing benchmarking resource via its homepage (\url{www.sdspeople.fudan.edu.cn/zhuangxiahai/0/mmwhs/}).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge