Weinan Song
A Collaborative Ensemble Framework for CTR Prediction
Nov 20, 2024



Abstract:Recent advances in foundation models have established scaling laws that enable the development of larger models to achieve enhanced performance, motivating extensive research into large-scale recommendation models. However, simply increasing the model size in recommendation systems, even with large amounts of data, does not always result in the expected performance improvements. In this paper, we propose a novel framework, Collaborative Ensemble Training Network (CETNet), to leverage multiple distinct models, each with its own embedding table, to capture unique feature interaction patterns. Unlike naive model scaling, our approach emphasizes diversity and collaboration through collaborative learning, where models iteratively refine their predictions. To dynamically balance contributions from each model, we introduce a confidence-based fusion mechanism using general softmax, where model confidence is computed via negation entropy. This design ensures that more confident models have a greater influence on the final prediction while benefiting from the complementary strengths of other models. We validate our framework on three public datasets (AmazonElectronics, TaobaoAds, and KuaiVideo) as well as a large-scale industrial dataset from Meta, demonstrating its superior performance over individual models and state-of-the-art baselines. Additionally, we conduct further experiments on the Criteo and Avazu datasets to compare our method with the multi-embedding paradigm. Our results show that our framework achieves comparable or better performance with smaller embedding sizes, offering a scalable and efficient solution for CTR prediction tasks.
Progressive Energy-Based Cooperative Learning for Multi-Domain Image-to-Image Translation
Jun 26, 2023Abstract:This paper studies a novel energy-based cooperative learning framework for multi-domain image-to-image translation. The framework consists of four components: descriptor, translator, style encoder, and style generator. The descriptor is a multi-head energy-based model that represents a multi-domain image distribution. The components of translator, style encoder, and style generator constitute a diversified image generator. Specifically, given an input image from a source domain, the translator turns it into a stylised output image of the target domain according to a style code, which can be inferred by the style encoder from a reference image or produced by the style generator from a random noise. Since the style generator is represented as an domain-specific distribution of style codes, the translator can provide a one-to-many transformation (i.e., diversified generation) between source domain and target domain. To train our framework, we propose a likelihood-based multi-domain cooperative learning algorithm to jointly train the multi-domain descriptor and the diversified image generator (including translator, style encoder, and style generator modules) via multi-domain MCMC teaching, in which the descriptor guides the diversified image generator to shift its probability density toward the data distribution, while the diversified image generator uses its randomly translated images to initialize the descriptor's Langevin dynamics process for efficient sampling.
Oral-NeXF: 3D Oral Reconstruction with Neural X-ray Field from Panoramic Imaging
Mar 21, 2023Abstract:3D reconstruction of medical images from 2D images has increasingly become a challenging research topic with the advanced development of deep learning methods. Previous work in 3D reconstruction from limited (generally one or two) X-ray images mainly relies on learning from paired 2D and 3D images. In 3D oral reconstruction from panoramic imaging, the model also relies on some prior individual information, such as the dental arch curve or voxel-wise annotations, to restore the curved shape of the mandible during reconstruction. These limitations have hindered the use of single X-ray tomography in clinical applications. To address these challenges, we propose a new model that relies solely on projection data, including imaging direction and projection image, during panoramic scans to reconstruct the 3D oral structure. Our model builds on the neural radiance field by introducing multi-head prediction, dynamic sampling, and adaptive rendering, which accommodates the projection process of panoramic X-ray in dental imaging. Compared to end-to-end learning methods, our method achieves state-of-the-art performance without requiring additional supervision or prior knowledge.
MDT-Net: Multi-domain Transfer by Perceptual Supervision for Unpaired Images in OCT Scan
Mar 12, 2022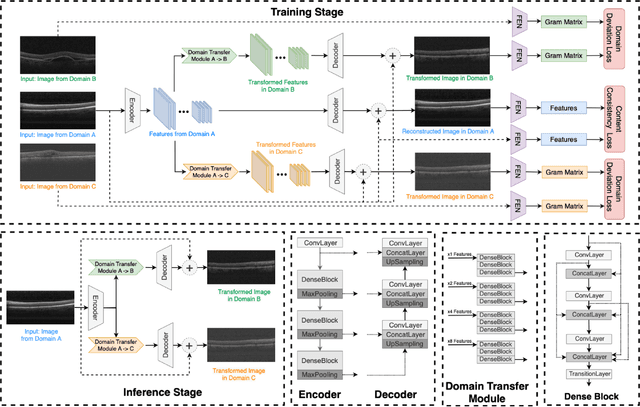
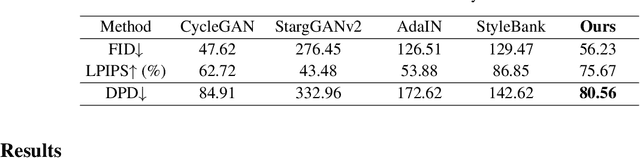
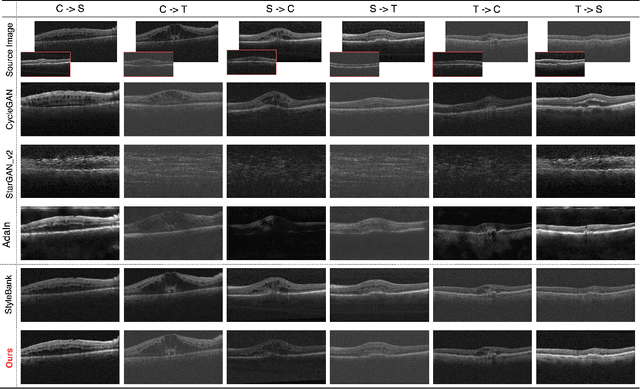
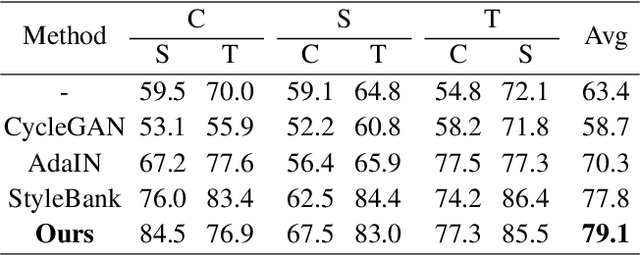
Abstract:Deep learning models tend to underperform in the presence of domain shifts. Domain transfer has recently emerged as a promising approach wherein images exhibiting a domain shift are transformed into other domains for augmentation or adaptation. However, with the absence of paired and annotated images, most domain transfer methods mainly rely on adversarial networks and weak cycle consistency, which could result in incomplete domain transfer or poor adherence to the original image content. In this paper, we introduce MDT-Net to address the limitations above through a multi-domain transfer model based on perceptual supervision. Specifically, our model consists of an encoder-decoder network, which aims to preserve anatomical structures, and multiple domain-specific transfer modules, which guide the domain transition through feature transformation. During the inference, MDT-Net can directly transfer images from the source domain to multiple target domains at one time without any reference image. To demonstrate the performance of MDT-Net, we evaluate it on RETOUCH dataset, comprising OCT scans from three different scanner devices (domains), for multi-domain transfer. We also take the transformed results as additional training images for fluid segmentation in OCT scans in the tasks of domain adaptation and data augmentation. Experimental results show that MDT-Net can outperform other domain transfer models qualitatively and quantitatively. Furthermore, the significant improvement in dice scores over multiple segmentation models also demonstrates the effectiveness and efficiency of our proposed method.
X2Teeth: 3D Teeth Reconstruction from a Single Panoramic Radiograph
Aug 30, 2021
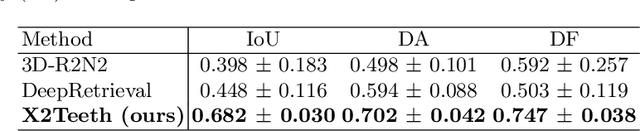


Abstract:3D teeth reconstruction from X-ray is important for dental diagnosis and many clinical operations. However, no existing work has explored the reconstruction of teeth for a whole cavity from a single panoramic radiograph. Different from single object reconstruction from photos, this task has the unique challenge of constructing multiple objects at high resolutions. To conquer this task, we develop a novel ConvNet X2Teeth that decomposes the task into teeth localization and single-shape estimation. We also introduce a patch-based training strategy, such that X2Teeth can be end-to-end trained for optimal performance. Extensive experiments show that our method can successfully estimate the 3D structure of the cavity and reflect the details for each tooth. Moreover, X2Teeth achieves a reconstruction IoU of 0.681, which significantly outperforms the encoder-decoder method by $1.71X and the retrieval-based method by $1.52X. Our method can also be promising for other multi-anatomy 3D reconstruction tasks.
Atlas-aware ConvNetfor Accurate yet Robust Anatomical Segmentation
Feb 02, 2021
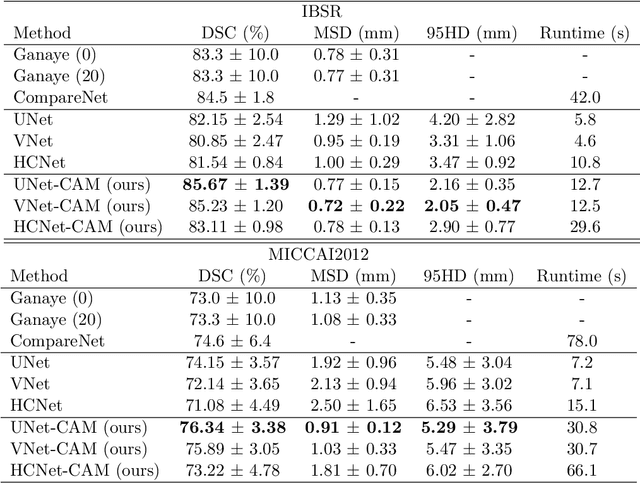
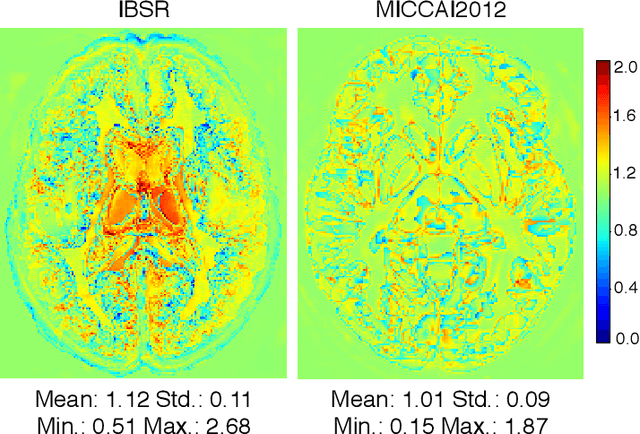
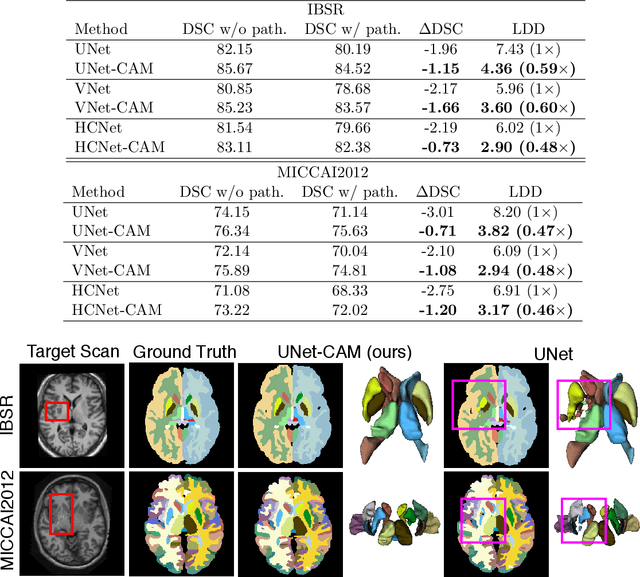
Abstract:Convolutional networks (ConvNets) have achieved promising accuracy for various anatomical segmentation tasks. Despite the success, these methods can be sensitive to data appearance variations. Considering the large variability of scans caused by artifacts, pathologies, and scanning setups, robust ConvNets are vital for clinical applications, while have not been fully explored. In this paper, we propose to mitigate the challenge by enabling ConvNets' awareness of the underlying anatomical invariances among imaging scans. Specifically, we introduce a fully convolutional Constraint Adoption Module (CAM) that incorporates probabilistic atlas priors as explicit constraints for predictions over a locally connected Conditional Random Field (CFR), which effectively reinforces the anatomical consistency of the labeling outputs. We design the CAM to be flexible for boosting various ConvNet, and compact for co-optimizing with ConvNets for fusion parameters that leads to the optimal performance. We show the advantage of such atlas priors fusion is two-fold with two brain parcellation tasks. First, our models achieve state-of-the-art accuracy among ConvNet-based methods on both datasets, by significantly reducing structural abnormalities of predictions. Second, we can largely boost the robustness of existing ConvNets, proved by: (i) testing on scans with synthetic pathologies, and (ii) training and evaluation on scans of different scanning setups across datasets. Our method is proposing to be easily adopted to existing ConvNets by fine-tuning with CAM plugged in for accuracy and robustness boosts.
Exploring Instance-Level Uncertainty for Medical Detection
Jan 09, 2021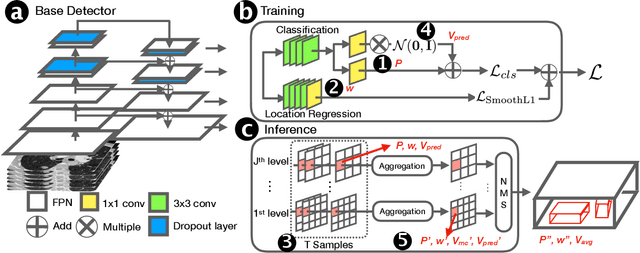
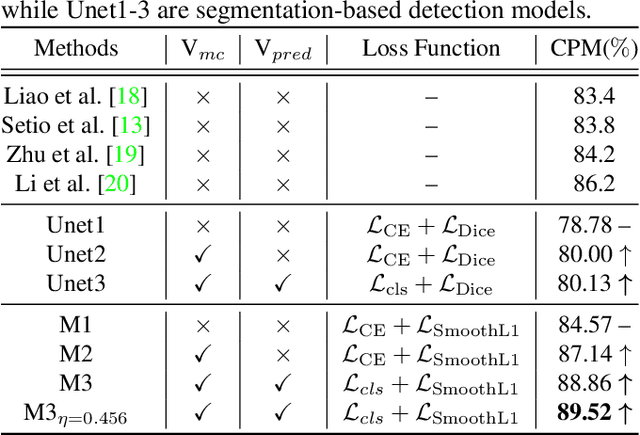
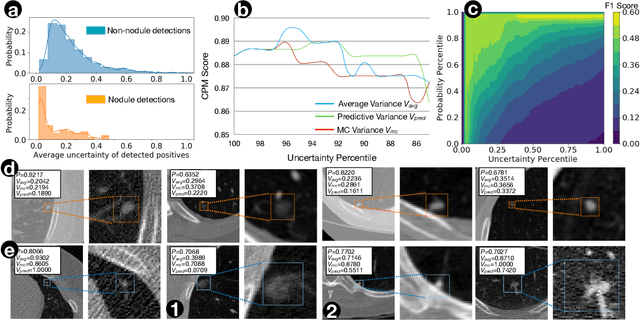
Abstract:The ability of deep learning to predict with uncertainty is recognized as key for its adoption in clinical routines. Moreover, performance gain has been enabled by modelling uncertainty according to empirical evidence. While previous work has widely discussed the uncertainty estimation in segmentation and classification tasks, its application on bounding-box-based detection has been limited, mainly due to the challenge of bounding box aligning. In this work, we explore to augment a 2.5D detection CNN with two different bounding-box-level (or instance-level) uncertainty estimates, i.e., predictive variance and Monte Carlo (MC) sample variance. Experiments are conducted for lung nodule detection on LUNA16 dataset, a task where significant semantic ambiguities can exist between nodules and non-nodules. Results show that our method improves the evaluating score from 84.57% to 88.86% by utilizing a combination of both types of variances. Moreover, we show the generated uncertainty enables superior operating points compared to using the probability threshold only, and can further boost the performance to 89.52%. Example nodule detections are visualized to further illustrate the advantages of our method.
Oral-3D: Reconstructing the 3D Bone Structure of Oral Cavity from 2D Panoramic X-ray
Mar 26, 2020


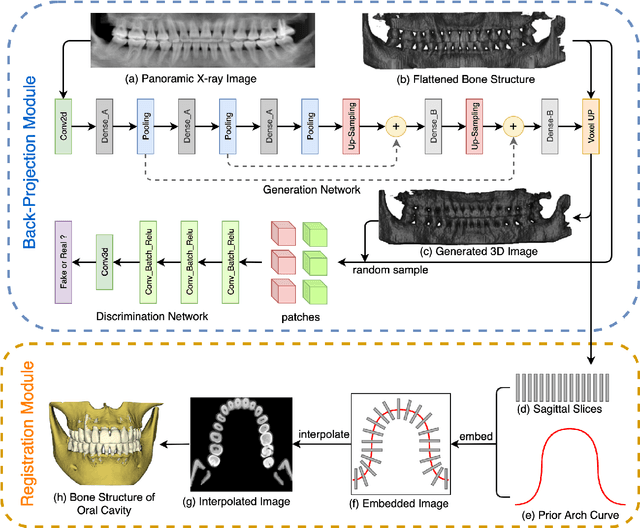
Abstract:Panoramic X-ray and Cone Beam Computed Tomography (CBCT) are two of the most general imaging methods in digital dentistry. While CBCT can provide higher-dimension information, the panoramic X-ray has the advantages of lower radiation dose and cost. Consequently, generating 3D information of bony tissues from the X-ray that can reflect dental diseases is of great interest. This technique can be even more helpful for developing areas where the CBCT is not always available due to the lack of screening machines or high screening cost. In this paper, we present \textit{Oral-3D} to reconstruct the bone structure of oral cavity from a single panoramic X-ray image by taking advantage of some prior knowledge in oral structure, which conventionally can only be obtained by a 3D imaging method like CBCT. Specifically, we first train a generative network to back project the 2D X-ray image into 3D space, then restore the bone structure by registering the generated 3D image with the prior shape of the dental arch. To be noted, \textit{Oral-3D} can restore both the density of bony tissues and the curved mandible surface. Experimental results show that our framework can reconstruct the 3D structure with significantly high quality. To the best of our knowledge, this is the first work that explores 3D reconstruction from a 2D image in dental health.
T-Net: A Template-Supervised Network for Task-specific Feature Extraction in Biomedical Image Analysis
Feb 19, 2020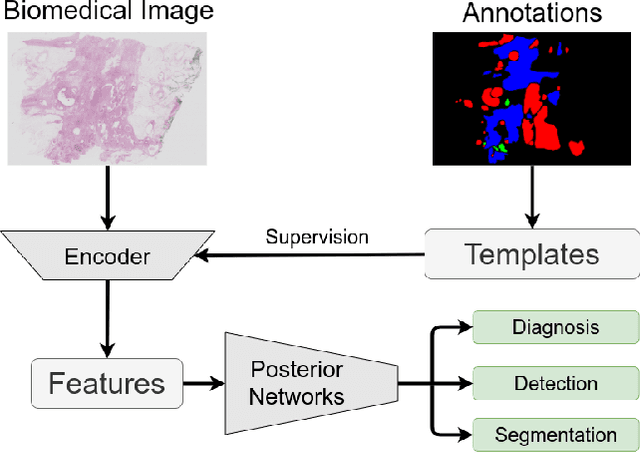
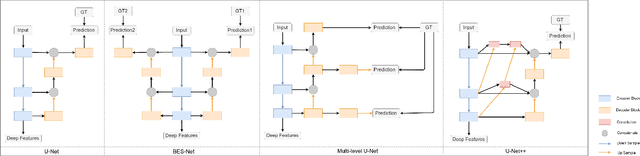

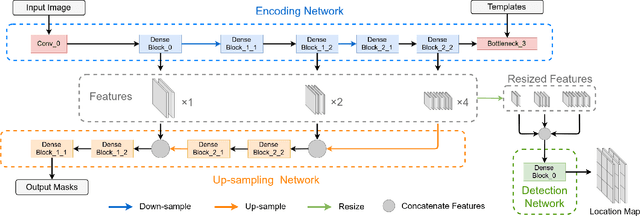
Abstract:Existing deep learning methods depend on an encoder-decoder structure to learn feature representation from the segmentation annotation in biomedical image analysis. However, the effectiveness of feature extraction under this structure decreases due to the indirect optimization process, limited training data size, and simplex supervision method. In this paper, we propose a template-supervised network T-Net for task-specific feature extraction. Specifically, we first obtain templates from pixel-level annotations by down-sampling binary masks of recognition targets according to specific tasks. Then, we directly train the encoding network under the supervision of the derived task-specific templates. Finally, we combine the resulting encoding network with a posterior network for the specific task, e.g. an up-sampling network for segmentation or a region proposal network for detection. Extensive experiments on three public datasets (BraTS-17, MoNuSeg and IDRiD) show that T-Net achieves competitive results to the state-of-the-art methods and superior performance to an encoder-decoder based network. To the best of our knowledge, this is the first in-depth study to improve feature extraction by directly supervise the encoding network and by applying task-specific supervision in biomedical image analysis.
CompareNet: Anatomical Segmentation Network with Deep Non-local Label Fusion
Oct 10, 2019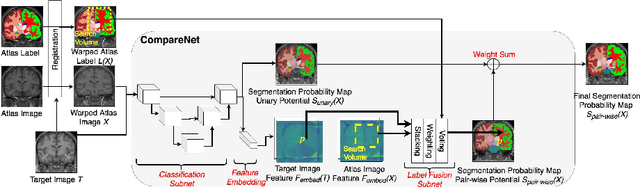
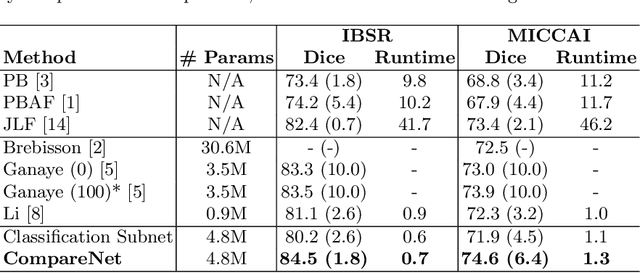
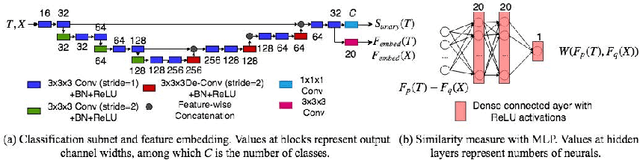

Abstract:Label propagation is a popular technique for anatomical segmentation. In this work, we propose a novel deep framework for label propagation based on non-local label fusion. Our framework, named CompareNet, incorporates subnets for both extracting discriminating features, and learning the similarity measure, which lead to accurate segmentation. We also introduce the voxel-wise classification as an unary potential to the label fusion function, for alleviating the search failure issue of the existing non-local fusion strategies. Moreover, CompareNet is end-to-end trainable, and all the parameters are learnt together for the optimal performance. By evaluating CompareNet on two public datasets IBSRv2 and MICCAI 2012 for brain segmentation, we show it outperforms state-of-the-art methods in accuracy, while being robust to pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge