Nima Tajbakhsh
Quantization-Aware Distillation for NVFP4 Inference Accuracy Recovery
Jan 27, 2026Abstract:This technical report presents quantization-aware distillation (QAD) and our best practices for recovering accuracy of NVFP4-quantized large language models (LLMs) and vision-language models (VLMs). QAD distills a full-precision teacher model into a quantized student model using a KL divergence loss. While applying distillation to quantized models is not a new idea, we observe key advantages of QAD for today's LLMs: 1. It shows remarkable effectiveness and stability for models trained through multi-stage post-training pipelines, including supervised fine-tuning (SFT), reinforcement learning (RL), and model merging, where traditional quantization-aware training (QAT) suffers from engineering complexity and training instability; 2. It is robust to data quality and coverage, enabling accuracy recovery without full training data. We evaluate QAD across multiple post-trained models including AceReason Nemotron, Nemotron 3 Nano, Nemotron Nano V2, Nemotron Nano V2 VL (VLM), and Llama Nemotron Super v1, showing consistent recovery to near-BF16 accuracy.
NVIDIA Nemotron 3: Efficient and Open Intelligence
Dec 24, 2025Abstract:We introduce the Nemotron 3 family of models - Nano, Super, and Ultra. These models deliver strong agentic, reasoning, and conversational capabilities. The Nemotron 3 family uses a Mixture-of-Experts hybrid Mamba-Transformer architecture to provide best-in-class throughput and context lengths of up to 1M tokens. Super and Ultra models are trained with NVFP4 and incorporate LatentMoE, a novel approach that improves model quality. The two larger models also include MTP layers for faster text generation. All Nemotron 3 models are post-trained using multi-environment reinforcement learning enabling reasoning, multi-step tool use, and support granular reasoning budget control. Nano, the smallest model, outperforms comparable models in accuracy while remaining extremely cost-efficient for inference. Super is optimized for collaborative agents and high-volume workloads such as IT ticket automation. Ultra, the largest model, provides state-of-the-art accuracy and reasoning performance. Nano is released together with its technical report and this white paper, while Super and Ultra will follow in the coming months. We will openly release the model weights, pre- and post-training software, recipes, and all data for which we hold redistribution rights.
Nemotron 3 Nano: Open, Efficient Mixture-of-Experts Hybrid Mamba-Transformer Model for Agentic Reasoning
Dec 23, 2025Abstract:We present Nemotron 3 Nano 30B-A3B, a Mixture-of-Experts hybrid Mamba-Transformer language model. Nemotron 3 Nano was pretrained on 25 trillion text tokens, including more than 3 trillion new unique tokens over Nemotron 2, followed by supervised fine tuning and large-scale RL on diverse environments. Nemotron 3 Nano achieves better accuracy than our previous generation Nemotron 2 Nano while activating less than half of the parameters per forward pass. It achieves up to 3.3x higher inference throughput than similarly-sized open models like GPT-OSS-20B and Qwen3-30B-A3B-Thinking-2507, while also being more accurate on popular benchmarks. Nemotron 3 Nano demonstrates enhanced agentic, reasoning, and chat abilities and supports context lengths up to 1M tokens. We release both our pretrained Nemotron 3 Nano 30B-A3B Base and post-trained Nemotron 3 Nano 30B-A3B checkpoints on Hugging Face.
NVIDIA Nemotron Nano 2: An Accurate and Efficient Hybrid Mamba-Transformer Reasoning Model
Aug 21, 2025



Abstract:We introduce Nemotron-Nano-9B-v2, a hybrid Mamba-Transformer language model designed to increase throughput for reasoning workloads while achieving state-of-the-art accuracy compared to similarly-sized models. Nemotron-Nano-9B-v2 builds on the Nemotron-H architecture, in which the majority of the self-attention layers in the common Transformer architecture are replaced with Mamba-2 layers, to achieve improved inference speed when generating the long thinking traces needed for reasoning. We create Nemotron-Nano-9B-v2 by first pre-training a 12-billion-parameter model (Nemotron-Nano-12B-v2-Base) on 20 trillion tokens using an FP8 training recipe. After aligning Nemotron-Nano-12B-v2-Base, we employ the Minitron strategy to compress and distill the model with the goal of enabling inference on up to 128k tokens on a single NVIDIA A10G GPU (22GiB of memory, bfloat16 precision). Compared to existing similarly-sized models (e.g., Qwen3-8B), we show that Nemotron-Nano-9B-v2 achieves on-par or better accuracy on reasoning benchmarks while achieving up to 6x higher inference throughput in reasoning settings like 8k input and 16k output tokens. We are releasing Nemotron-Nano-9B-v2, Nemotron-Nano12B-v2-Base, and Nemotron-Nano-9B-v2-Base checkpoints along with the majority of our pre- and post-training datasets on Hugging Face.
Efficient Hybrid Language Model Compression through Group-Aware SSM Pruning
Apr 15, 2025Abstract:Hybrid LLM architectures that combine Attention and State Space Models (SSMs) achieve state-of-the-art accuracy and runtime performance. Recent work has demonstrated that applying compression and distillation to Attention-only models yields smaller, more accurate models at a fraction of the training cost. In this work, we explore the effectiveness of compressing Hybrid architectures. We introduce a novel group-aware pruning strategy that preserves the structural integrity of SSM blocks and their sequence modeling capabilities. Furthermore, we demonstrate the necessity of such SSM pruning to achieve improved accuracy and inference speed compared to traditional approaches. Our compression recipe combines SSM, FFN, embedding dimension, and layer pruning, followed by knowledge distillation-based retraining, similar to the MINITRON technique. Using this approach, we compress the Nemotron-H 8B Hybrid model down to 4B parameters with up to 40x fewer training tokens. The resulting model surpasses the accuracy of similarly-sized models while achieving 2x faster inference, significantly advancing the Pareto frontier.
Nemotron-H: A Family of Accurate and Efficient Hybrid Mamba-Transformer Models
Apr 10, 2025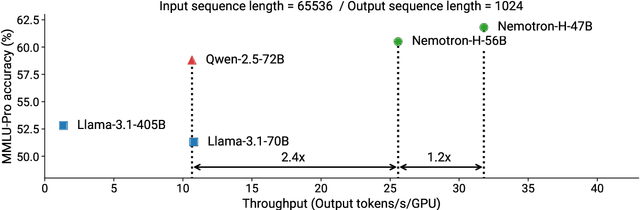

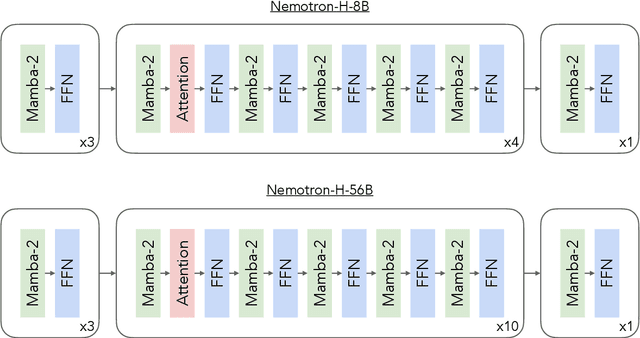
Abstract:As inference-time scaling becomes critical for enhanced reasoning capabilities, it is increasingly becoming important to build models that are efficient to infer. We introduce Nemotron-H, a family of 8B and 56B/47B hybrid Mamba-Transformer models designed to reduce inference cost for a given accuracy level. To achieve this goal, we replace the majority of self-attention layers in the common Transformer model architecture with Mamba layers that perform constant computation and require constant memory per generated token. We show that Nemotron-H models offer either better or on-par accuracy compared to other similarly-sized state-of-the-art open-sourced Transformer models (e.g., Qwen-2.5-7B/72B and Llama-3.1-8B/70B), while being up to 3$\times$ faster at inference. To further increase inference speed and reduce the memory required at inference time, we created Nemotron-H-47B-Base from the 56B model using a new compression via pruning and distillation technique called MiniPuzzle. Nemotron-H-47B-Base achieves similar accuracy to the 56B model, but is 20% faster to infer. In addition, we introduce an FP8-based training recipe and show that it can achieve on par results with BF16-based training. This recipe is used to train the 56B model. All Nemotron-H models will be released, with support in Hugging Face, NeMo, and Megatron-LM.
Training Video Foundation Models with NVIDIA NeMo
Mar 17, 2025Abstract:Video Foundation Models (VFMs) have recently been used to simulate the real world to train physical AI systems and develop creative visual experiences. However, there are significant challenges in training large-scale, high quality VFMs that can generate high-quality videos. We present a scalable, open-source VFM training pipeline with NVIDIA NeMo, providing accelerated video dataset curation, multimodal data loading, and parallelized video diffusion model training and inference. We also provide a comprehensive performance analysis highlighting best practices for efficient VFM training and inference.
Llama 3 Meets MoE: Efficient Upcycling
Dec 13, 2024



Abstract:Scaling large language models (LLMs) significantly improves performance but comes with prohibitive computational costs. Mixture-of-Experts (MoE) models offer an efficient alternative, increasing capacity without a proportional rise in compute requirements. However, training MoE models from scratch poses challenges like overfitting and routing instability. We present an efficient training recipe leveraging pre-trained dense checkpoints, training an 8-Expert Top-2 MoE model from Llama 3-8B with less than $1\%$ of typical pre-training compute. Our approach enhances downstream performance on academic benchmarks, achieving a $\textbf{2%}$ improvement in 0-shot accuracy on MMLU, while reaching a Model FLOPs Utilization (MFU) of $\textbf{46.8%}$ during training using our framework. We also integrate online upcycling in NeMo for seamless use of pre-trained weights, enabling cost-effective development of high-capacity MoE models.
Elucidating Optimal Reward-Diversity Tradeoffs in Text-to-Image Diffusion Models
Sep 09, 2024Abstract:Text-to-image (T2I) diffusion models have become prominent tools for generating high-fidelity images from text prompts. However, when trained on unfiltered internet data, these models can produce unsafe, incorrect, or stylistically undesirable images that are not aligned with human preferences. To address this, recent approaches have incorporated human preference datasets to fine-tune T2I models or to optimize reward functions that capture these preferences. Although effective, these methods are vulnerable to reward hacking, where the model overfits to the reward function, leading to a loss of diversity in the generated images. In this paper, we prove the inevitability of reward hacking and study natural regularization techniques like KL divergence and LoRA scaling, and their limitations for diffusion models. We also introduce Annealed Importance Guidance (AIG), an inference-time regularization inspired by Annealed Importance Sampling, which retains the diversity of the base model while achieving Pareto-Optimal reward-diversity tradeoffs. Our experiments demonstrate the benefits of AIG for Stable Diffusion models, striking the optimal balance between reward optimization and image diversity. Furthermore, a user study confirms that AIG improves diversity and quality of generated images across different model architectures and reward functions.
StarCoder 2 and The Stack v2: The Next Generation
Feb 29, 2024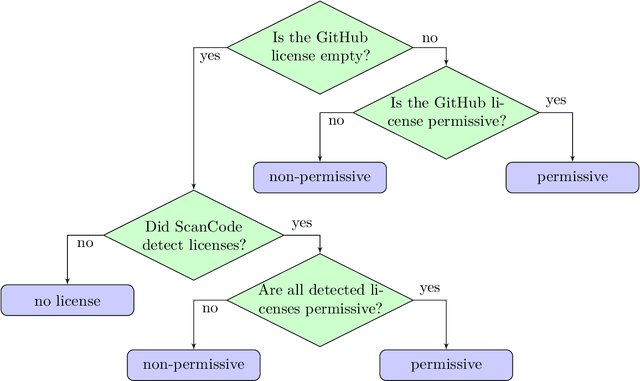
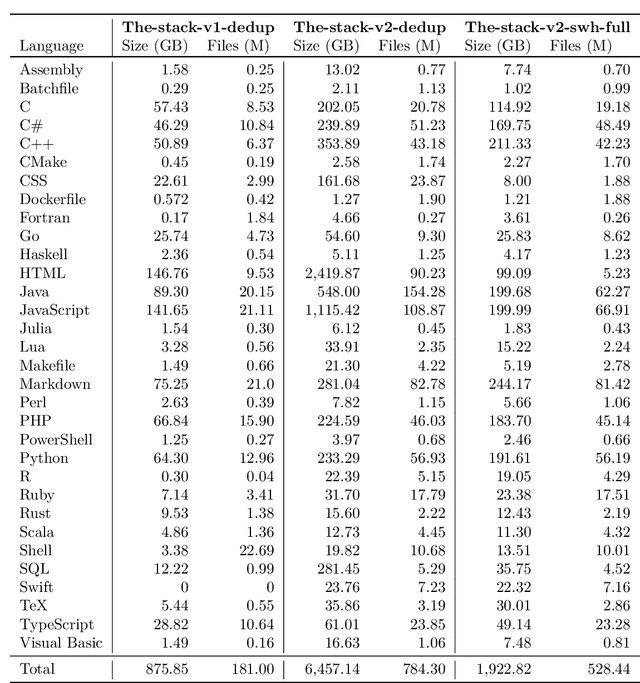
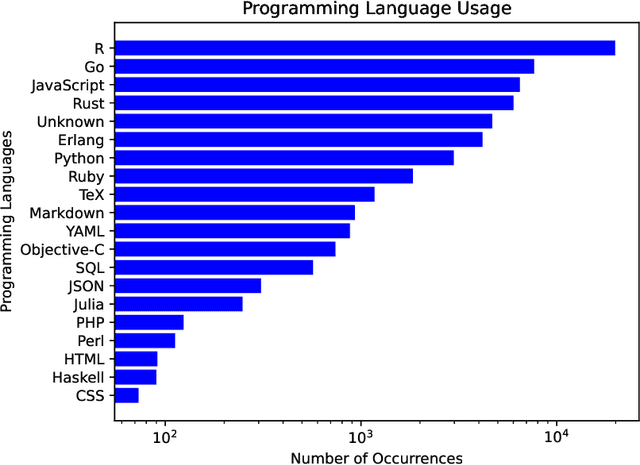

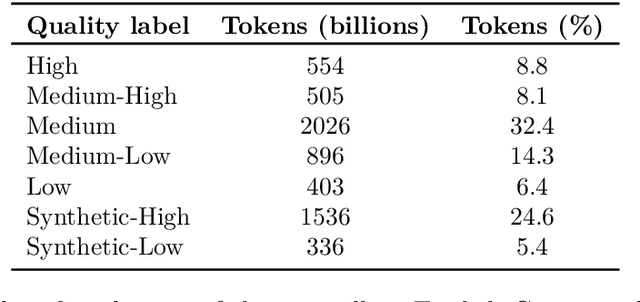
Abstract:The BigCode project, an open-scientific collaboration focused on the responsible development of Large Language Models for Code (Code LLMs), introduces StarCoder2. In partnership with Software Heritage (SWH), we build The Stack v2 on top of the digital commons of their source code archive. Alongside the SWH repositories spanning 619 programming languages, we carefully select other high-quality data sources, such as GitHub pull requests, Kaggle notebooks, and code documentation. This results in a training set that is 4x larger than the first StarCoder dataset. We train StarCoder2 models with 3B, 7B, and 15B parameters on 3.3 to 4.3 trillion tokens and thoroughly evaluate them on a comprehensive set of Code LLM benchmarks. We find that our small model, StarCoder2-3B, outperforms other Code LLMs of similar size on most benchmarks, and also outperforms StarCoderBase-15B. Our large model, StarCoder2- 15B, significantly outperforms other models of comparable size. In addition, it matches or outperforms CodeLlama-34B, a model more than twice its size. Although DeepSeekCoder- 33B is the best-performing model at code completion for high-resource languages, we find that StarCoder2-15B outperforms it on math and code reasoning benchmarks, as well as several low-resource languages. We make the model weights available under an OpenRAIL license and ensure full transparency regarding the training data by releasing the SoftWare Heritage persistent IDentifiers (SWHIDs) of the source code data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge