Tianqi Wu
FlashOverlap: A Lightweight Design for Efficiently Overlapping Communication and Computation
Apr 28, 2025Abstract:Generative models have achieved remarkable success across various applications, driving the demand for multi-GPU computing. Inter-GPU communication becomes a bottleneck in multi-GPU computing systems, particularly on consumer-grade GPUs. By exploiting concurrent hardware execution, overlapping computation and communication latency is an effective technique for mitigating the communication overhead. We identify that an efficient and adaptable overlapping design should satisfy (1) tile-wise overlapping to maximize the overlapping opportunity, (2) interference-free computation to maintain the original computational performance, and (3) communication agnosticism to reduce the development burden against varying communication primitives. Nevertheless, current designs fail to simultaneously optimize for all of those features. To address the issue, we propose FlashOverlap, a lightweight design characterized by tile-wise overlapping, interference-free computation, and communication agnosticism. FlashOverlap utilizes a novel signaling mechanism to identify tile-wise data dependency without interrupting the computation process, and reorders data to contiguous addresses, enabling communication by simply calling NCCL APIs. Experiments show that such a lightweight design achieves up to 1.65x speedup, outperforming existing works in most cases.
MoA: Mixture of Sparse Attention for Automatic Large Language Model Compression
Jun 21, 2024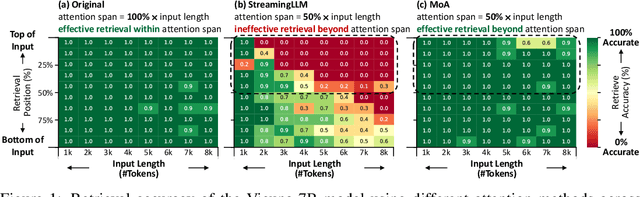
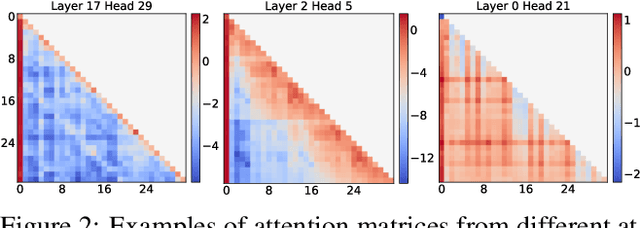

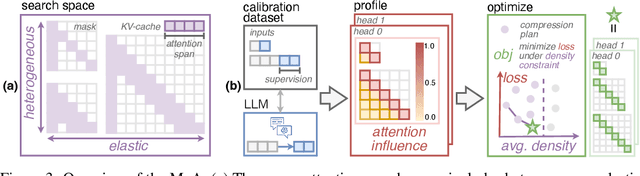
Abstract:Sparse attention can effectively mitigate the significant memory and throughput demands of Large Language Models (LLMs) in long contexts. Existing methods typically employ a uniform sparse attention mask, applying the same sparse pattern across different attention heads and input lengths. However, this uniform approach fails to capture the diverse attention patterns inherent in LLMs, ignoring their distinct accuracy-latency trade-offs. To address this challenge, we propose the Mixture of Attention (MoA), which automatically tailors distinct sparse attention configurations to different heads and layers. MoA constructs and navigates a search space of various attention patterns and their scaling rules relative to input sequence lengths. It profiles the model, evaluates potential configurations, and pinpoints the optimal sparse attention compression plan. MoA adapts to varying input sizes, revealing that some attention heads expand their focus to accommodate longer sequences, while other heads consistently concentrate on fixed-length local contexts. Experiments show that MoA increases the effective context length by $3.9\times$ with the same average attention span, boosting retrieval accuracy by $1.5-7.1\times$ over the uniform-attention baseline across Vicuna-7B, Vicuna-13B, and Llama3-8B models. Moreover, MoA narrows the capability gaps between sparse and dense models, reducing the maximum relative performance drop from $9\%-36\%$ to within $5\%$ across two long-context understanding benchmarks. MoA achieves a $1.2-1.4\times$ GPU memory reduction and boosts decode throughput by $5.5-6.7 \times$ for 7B and 13B dense models on a single GPU, with minimal impact on performance.
EGR: Equivariant Graph Refinement and Assessment of 3D Protein Complex Structures
May 24, 2022



Abstract:Protein complexes are macromolecules essential to the functioning and well-being of all living organisms. As the structure of a protein complex, in particular its region of interaction between multiple protein subunits (i.e., chains), has a notable influence on the biological function of the complex, computational methods that can quickly and effectively be used to refine and assess the quality of a protein complex's 3D structure can directly be used within a drug discovery pipeline to accelerate the development of new therapeutics and improve the efficacy of future vaccines. In this work, we introduce the Equivariant Graph Refiner (EGR), a novel E(3)-equivariant graph neural network (GNN) for multi-task structure refinement and assessment of protein complexes. Our experiments on new, diverse protein complex datasets, all of which we make publicly available in this work, demonstrate the state-of-the-art effectiveness of EGR for atomistic refinement and assessment of protein complexes and outline directions for future work in the field. In doing so, we establish a baseline for future studies in macromolecular refinement and structure analysis.
What is Learned in Knowledge Graph Embeddings?
Oct 19, 2021
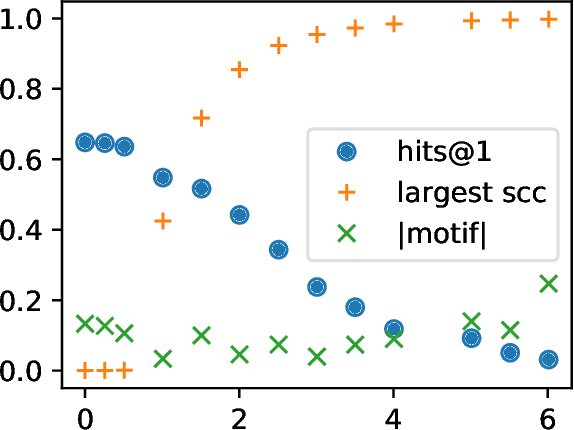
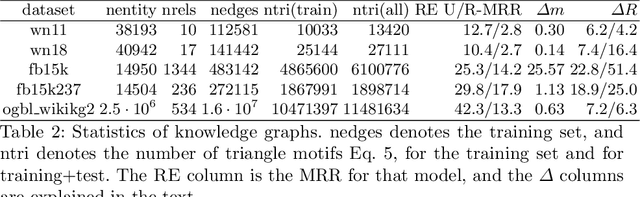

Abstract:A knowledge graph (KG) is a data structure which represents entities and relations as the vertices and edges of a directed graph with edge types. KGs are an important primitive in modern machine learning and artificial intelligence. Embedding-based models, such as the seminal TransE [Bordes et al., 2013] and the recent PairRE [Chao et al., 2020] are among the most popular and successful approaches for representing KGs and inferring missing edges (link completion). Their relative success is often credited in the literature to their ability to learn logical rules between the relations. In this work, we investigate whether learning rules between relations is indeed what drives the performance of embedding-based methods. We define motif learning and two alternative mechanisms, network learning (based only on the connectivity of the KG, ignoring the relation types), and unstructured statistical learning (ignoring the connectivity of the graph). Using experiments on synthetic KGs, we show that KG models can learn motifs and how this ability is degraded by non-motif (noise) edges. We propose tests to distinguish the contributions of the three mechanisms to performance, and apply them to popular KG benchmarks. We also discuss an issue with the standard performance testing protocol and suggest an improvement. To appear in the proceedings of Complex Networks 2021.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge