Paul M. Thompson
for the Alzheimer's Disease Neuroimaging Initiative
Deep Models, Shallow Alignment: Uncovering the Granularity Mismatch in Neural Decoding
Jan 29, 2026Abstract:Neural visual decoding is a central problem in brain computer interface research, aiming to reconstruct human visual perception and to elucidate the structure of neural representations. However, existing approaches overlook a fundamental granularity mismatch between human and machine vision, where deep vision models emphasize semantic invariance by suppressing local texture information, whereas neural signals preserve an intricate mixture of low-level visual attributes and high-level semantic content. To address this mismatch, we propose Shallow Alignment, a novel contrastive learning strategy that aligns neural signals with intermediate representations of visual encoders rather than their final outputs, thereby striking a better balance between low-level texture details and high-level semantic features. Extensive experiments across multiple benchmarks demonstrate that Shallow Alignment significantly outperforms standard final-layer alignment, with performance gains ranging from 22% to 58% across diverse vision backbones. Notably, our approach effectively unlocks the scaling law in neural visual decoding, enabling decoding performance to scale predictably with the capacity of pre-trained vision backbones. We further conduct systematic empirical analyses to shed light on the mechanisms underlying the observed performance gains.
Multi-modal Imputation for Alzheimer's Disease Classification
Jan 28, 2026Abstract:Deep learning has been successful in predicting neurodegenerative disorders, such as Alzheimer's disease, from magnetic resonance imaging (MRI). Combining multiple imaging modalities, such as T1-weighted (T1) and diffusion-weighted imaging (DWI) scans, can increase diagnostic performance. However, complete multimodal datasets are not always available. We use a conditional denoising diffusion probabilistic model to impute missing DWI scans from T1 scans. We perform extensive experiments to evaluate whether such imputation improves the accuracy of uni-modal and bi-modal deep learning models for 3-way Alzheimer's disease classification-cognitively normal, mild cognitive impairment, and Alzheimer's disease. We observe improvements in several metrics, particularly those sensitive to minority classes, for several imputation configurations.
How Much Data Is Enough? Uniform Convergence Bounds for Generative & Vision-Language Models under Low-Dimensional Structure
Dec 28, 2025Abstract:Modern generative and vision-language models (VLMs) are increasingly used in scientific and medical decision support, where predicted probabilities must be both accurate and well calibrated. Despite strong empirical results with moderate data, it remains unclear when such predictions generalize uniformly across inputs, classes, or subpopulations, rather than only on average-a critical issue in biomedicine, where rare conditions and specific groups can exhibit large errors even when overall loss is low. We study this question from a finite-sample perspective and ask: under what structural assumptions can generative and VLM-based predictors achieve uniformly accurate and calibrated behavior with practical sample sizes? Rather than analyzing arbitrary parameterizations, we focus on induced families of classifiers obtained by varying prompts or semantic embeddings within a restricted representation space. When model outputs depend smoothly on a low-dimensional semantic representation-an assumption supported by spectral structure in text and joint image-text embeddings-classical uniform convergence tools yield meaningful non-asymptotic guarantees. Our main results give finite-sample uniform convergence bounds for accuracy and calibration functionals of VLM-induced classifiers under Lipschitz stability with respect to prompt embeddings. The implied sample complexity depends on intrinsic/effective dimension, not ambient embedding dimension, and we further derive spectrum-dependent bounds that make explicit how eigenvalue decay governs data requirements. We conclude with implications for data-limited biomedical modeling, including when current dataset sizes can support uniformly reliable predictions and why average calibration metrics may miss worst-case miscalibration.
R-GenIMA: Integrating Neuroimaging and Genetics with Interpretable Multimodal AI for Alzheimer's Disease Progression
Dec 22, 2025Abstract:Early detection of Alzheimer's disease (AD) requires models capable of integrating macro-scale neuroanatomical alterations with micro-scale genetic susceptibility, yet existing multimodal approaches struggle to align these heterogeneous signals. We introduce R-GenIMA, an interpretable multimodal large language model that couples a novel ROI-wise vision transformer with genetic prompting to jointly model structural MRI and single nucleotide polymorphisms (SNPs) variations. By representing each anatomically parcellated brain region as a visual token and encoding SNP profiles as structured text, the framework enables cross-modal attention that links regional atrophy patterns to underlying genetic factors. Applied to the ADNI cohort, R-GenIMA achieves state-of-the-art performance in four-way classification across normal cognition (NC), subjective memory concerns (SMC), mild cognitive impairment (MCI), and AD. Beyond predictive accuracy, the model yields biologically meaningful explanations by identifying stage-specific brain regions and gene signatures, as well as coherent ROI-Gene association patterns across the disease continuum. Attention-based attribution revealed genes consistently enriched for established GWAS-supported AD risk loci, including APOE, BIN1, CLU, and RBFOX1. Stage-resolved neuroanatomical signatures identified shared vulnerability hubs across disease stages alongside stage-specific patterns: striatal involvement in subjective decline, frontotemporal engagement during prodromal impairment, and consolidated multimodal network disruption in AD. These results demonstrate that interpretable multimodal AI can synthesize imaging and genetics to reveal mechanistic insights, providing a foundation for clinically deployable tools that enable earlier risk stratification and inform precision therapeutic strategies in Alzheimer's disease.
Diffusion Bridge Models for 3D Medical Image Translation
Apr 21, 2025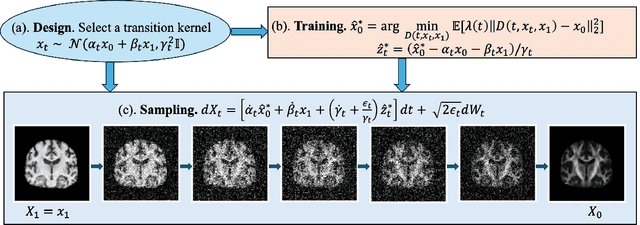
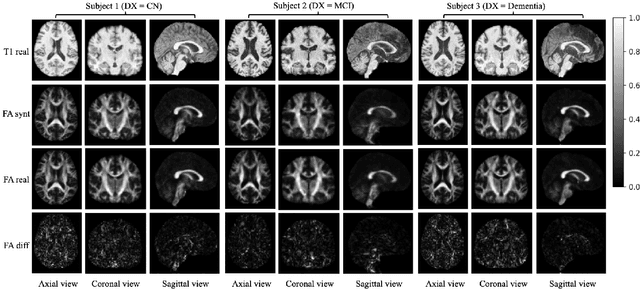
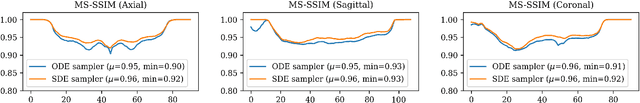
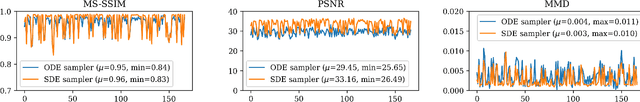
Abstract:Diffusion tensor imaging (DTI) provides crucial insights into the microstructure of the human brain, but it can be time-consuming to acquire compared to more readily available T1-weighted (T1w) magnetic resonance imaging (MRI). To address this challenge, we propose a diffusion bridge model for 3D brain image translation between T1w MRI and DTI modalities. Our model learns to generate high-quality DTI fractional anisotropy (FA) images from T1w images and vice versa, enabling cross-modality data augmentation and reducing the need for extensive DTI acquisition. We evaluate our approach using perceptual similarity, pixel-level agreement, and distributional consistency metrics, demonstrating strong performance in capturing anatomical structures and preserving information on white matter integrity. The practical utility of the synthetic data is validated through sex classification and Alzheimer's disease classification tasks, where the generated images achieve comparable performance to real data. Our diffusion bridge model offers a promising solution for improving neuroimaging datasets and supporting clinical decision-making, with the potential to significantly impact neuroimaging research and clinical practice.
MICCAI-CDMRI 2023 QuantConn Challenge Findings on Achieving Robust Quantitative Connectivity through Harmonized Preprocessing of Diffusion MRI
Nov 14, 2024



Abstract:White matter alterations are increasingly implicated in neurological diseases and their progression. International-scale studies use diffusion-weighted magnetic resonance imaging (DW-MRI) to qualitatively identify changes in white matter microstructure and connectivity. Yet, quantitative analysis of DW-MRI data is hindered by inconsistencies stemming from varying acquisition protocols. There is a pressing need to harmonize the preprocessing of DW-MRI datasets to ensure the derivation of robust quantitative diffusion metrics across acquisitions. In the MICCAI-CDMRI 2023 QuantConn challenge, participants were provided raw data from the same individuals collected on the same scanner but with two different acquisitions and tasked with preprocessing the DW-MRI to minimize acquisition differences while retaining biological variation. Submissions are evaluated on the reproducibility and comparability of cross-acquisition bundle-wise microstructure measures, bundle shape features, and connectomics. The key innovations of the QuantConn challenge are that (1) we assess bundles and tractography in the context of harmonization for the first time, (2) we assess connectomics in the context of harmonization for the first time, and (3) we have 10x additional subjects over prior harmonization challenge, MUSHAC and 100x over SuperMUDI. We find that bundle surface area, fractional anisotropy, connectome assortativity, betweenness centrality, edge count, modularity, nodal strength, and participation coefficient measures are most biased by acquisition and that machine learning voxel-wise correction, RISH mapping, and NeSH methods effectively reduce these biases. In addition, microstructure measures AD, MD, RD, bundle length, connectome density, efficiency, and path length are least biased by these acquisition differences.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024/019
DenseNet and Support Vector Machine classifications of major depressive disorder using vertex-wise cortical features
Nov 18, 2023



Abstract:Major depressive disorder (MDD) is a complex psychiatric disorder that affects the lives of hundreds of millions of individuals around the globe. Even today, researchers debate if morphological alterations in the brain are linked to MDD, likely due to the heterogeneity of this disorder. The application of deep learning tools to neuroimaging data, capable of capturing complex non-linear patterns, has the potential to provide diagnostic and predictive biomarkers for MDD. However, previous attempts to demarcate MDD patients and healthy controls (HC) based on segmented cortical features via linear machine learning approaches have reported low accuracies. In this study, we used globally representative data from the ENIGMA-MDD working group containing an extensive sample of people with MDD (N=2,772) and HC (N=4,240), which allows a comprehensive analysis with generalizable results. Based on the hypothesis that integration of vertex-wise cortical features can improve classification performance, we evaluated the classification of a DenseNet and a Support Vector Machine (SVM), with the expectation that the former would outperform the latter. As we analyzed a multi-site sample, we additionally applied the ComBat harmonization tool to remove potential nuisance effects of site. We found that both classifiers exhibited close to chance performance (balanced accuracy DenseNet: 51%; SVM: 53%), when estimated on unseen sites. Slightly higher classification performance (balanced accuracy DenseNet: 58%; SVM: 55%) was found when the cross-validation folds contained subjects from all sites, indicating site effect. In conclusion, the integration of vertex-wise morphometric features and the use of the non-linear classifier did not lead to the differentiability between MDD and HC. Our results support the notion that MDD classification on this combination of features and classifiers is unfeasible.
Video and Synthetic MRI Pre-training of 3D Vision Architectures for Neuroimage Analysis
Sep 09, 2023Abstract:Transfer learning represents a recent paradigm shift in the way we build artificial intelligence (AI) systems. In contrast to training task-specific models, transfer learning involves pre-training deep learning models on a large corpus of data and minimally fine-tuning them for adaptation to specific tasks. Even so, for 3D medical imaging tasks, we do not know if it is best to pre-train models on natural images, medical images, or even synthetically generated MRI scans or video data. To evaluate these alternatives, here we benchmarked vision transformers (ViTs) and convolutional neural networks (CNNs), initialized with varied upstream pre-training approaches. These methods were then adapted to three unique downstream neuroimaging tasks with a range of difficulty: Alzheimer's disease (AD) and Parkinson's disease (PD) classification, "brain age" prediction. Experimental tests led to the following key observations: 1. Pre-training improved performance across all tasks including a boost of 7.4% for AD classification and 4.6% for PD classification for the ViT and 19.1% for PD classification and reduction in brain age prediction error by 1.26 years for CNNs, 2. Pre-training on large-scale video or synthetic MRI data boosted performance of ViTs, 3. CNNs were robust in limited-data settings, and in-domain pretraining enhanced their performances, 4. Pre-training improved generalization to out-of-distribution datasets and sites. Overall, we benchmarked different vision architectures, revealing the value of pre-training them with emerging datasets for model initialization. The resulting pre-trained models can be adapted to a range of downstream neuroimaging tasks, even when training data for the target task is limited.
Incomplete Multimodal Learning for Complex Brain Disorders Prediction
May 25, 2023

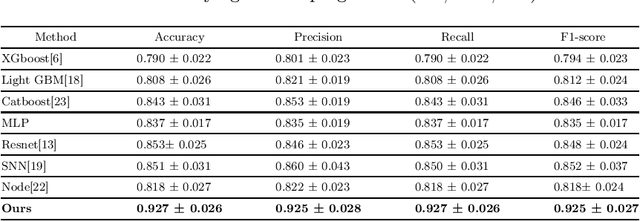

Abstract:Recent advancements in the acquisition of various brain data sources have created new opportunities for integrating multimodal brain data to assist in early detection of complex brain disorders. However, current data integration approaches typically need a complete set of biomedical data modalities, which may not always be feasible, as some modalities are only available in large-scale research cohorts and are prohibitive to collect in routine clinical practice. Especially in studies of brain diseases, research cohorts may include both neuroimaging data and genetic data, but for practical clinical diagnosis, we often need to make disease predictions only based on neuroimages. As a result, it is desired to design machine learning models which can use all available data (different data could provide complementary information) during training but conduct inference using only the most common data modality. We propose a new incomplete multimodal data integration approach that employs transformers and generative adversarial networks to effectively exploit auxiliary modalities available during training in order to improve the performance of a unimodal model at inference. We apply our new method to predict cognitive degeneration and disease outcomes using the multimodal imaging genetic data from Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort. Experimental results demonstrate that our approach outperforms the related machine learning and deep learning methods by a significant margin.
A Surface-Based Federated Chow Test Model for Integrating APOE Status, Tau Deposition Measure, and Hippocampal Surface Morphometry
Mar 31, 2023Abstract:Background: Alzheimer's Disease (AD) is the most common type of age-related dementia, affecting 6.2 million people aged 65 or older according to CDC data. It is commonly agreed that discovering an effective AD diagnosis biomarker could have enormous public health benefits, potentially preventing or delaying up to 40% of dementia cases. Tau neurofibrillary tangles are the primary driver of downstream neurodegeneration and subsequent cognitive impairment in AD, resulting in structural deformations such as hippocampal atrophy that can be observed in magnetic resonance imaging (MRI) scans. Objective: To build a surface-based model to 1) detect differences between APOE subgroups in patterns of tau deposition and hippocampal atrophy, and 2) use the extracted surface-based features to predict cognitive decline. Methods: Using data obtained from different institutions, we develop a surface-based federated Chow test model to study the synergistic effects of APOE, a previously reported significant risk factor of AD, and tau on hippocampal surface morphometry. Results: We illustrate that the APOE-specific morphometry features correlate with AD progression and better predict future AD conversion than other MRI biomarkers. For example, a strong association between atrophy and abnormal tau was identified in hippocampal subregion cornu ammonis 1 (CA1 subfield) and subiculum in e4 homozygote cohort. Conclusion: Our model allows for identifying MRI biomarkers for AD and cognitive decline prediction and may uncover a corner of the neural mechanism of the influence of APOE and tau deposition on hippocampal morphology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge