Nikhil Dhinagar
ADNI
Transferring Models Trained on Natural Images to 3D MRI via Position Encoded Slice Models
Mar 02, 2023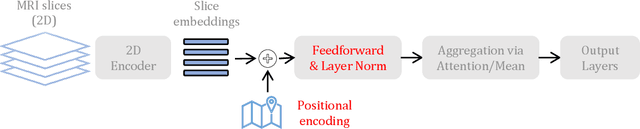
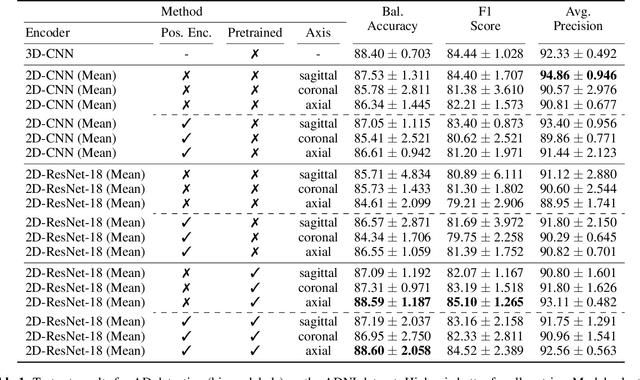
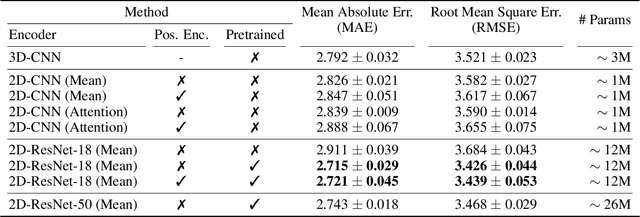
Abstract:Transfer learning has remarkably improved computer vision. These advances also promise improvements in neuroimaging, where training set sizes are often small. However, various difficulties arise in directly applying models pretrained on natural images to radiologic images, such as MRIs. In particular, a mismatch in the input space (2D images vs. 3D MRIs) restricts the direct transfer of models, often forcing us to consider only a few MRI slices as input. To this end, we leverage the 2D-Slice-CNN architecture of Gupta et al. (2021), which embeds all the MRI slices with 2D encoders (neural networks that take 2D image input) and combines them via permutation-invariant layers. With the insight that the pretrained model can serve as the 2D encoder, we initialize the 2D encoder with ImageNet pretrained weights that outperform those initialized and trained from scratch on two neuroimaging tasks -- brain age prediction on the UK Biobank dataset and Alzheimer's disease detection on the ADNI dataset. Further, we improve the modeling capabilities of 2D-Slice models by incorporating spatial information through position embeddings, which can improve the performance in some cases.
Towards Sparsified Federated Neuroimaging Models via Weight Pruning
Aug 24, 2022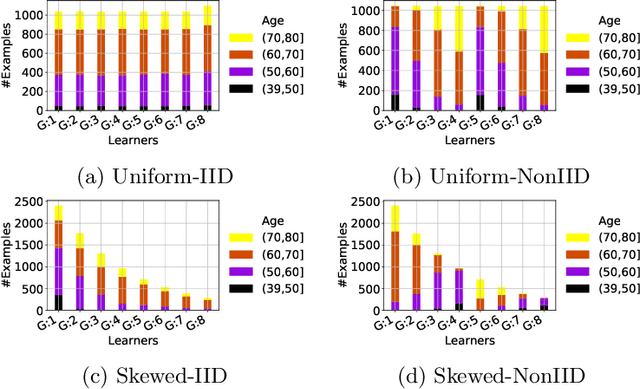
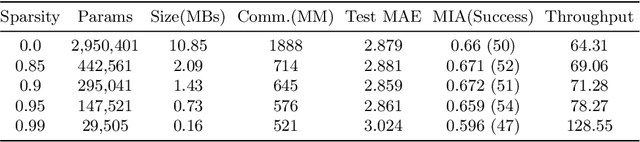
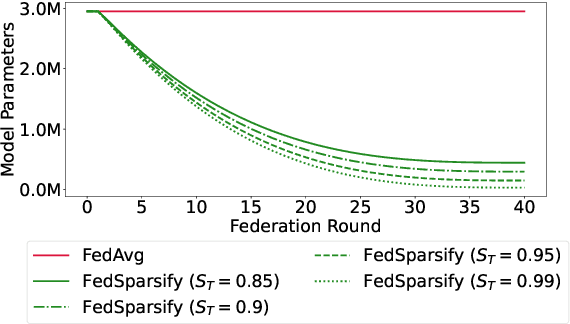
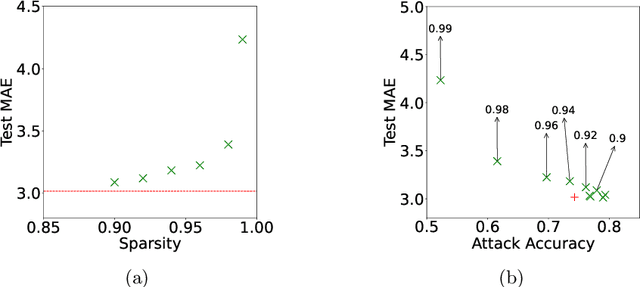
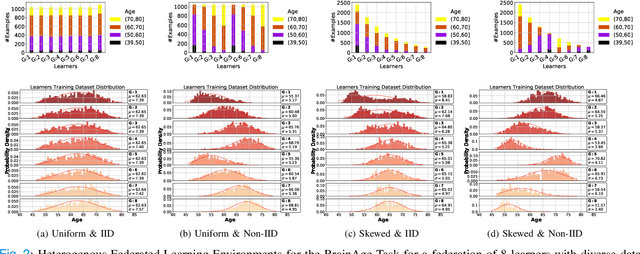
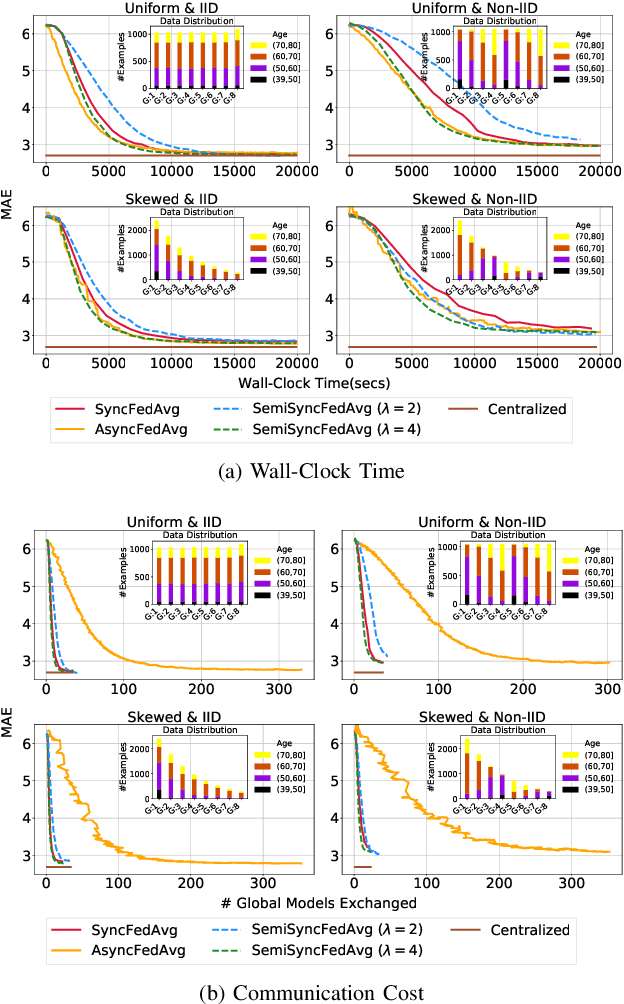
Abstract:Federated training of large deep neural networks can often be restrictive due to the increasing costs of communicating the updates with increasing model sizes. Various model pruning techniques have been designed in centralized settings to reduce inference times. Combining centralized pruning techniques with federated training seems intuitive for reducing communication costs -- by pruning the model parameters right before the communication step. Moreover, such a progressive model pruning approach during training can also reduce training times/costs. To this end, we propose FedSparsify, which performs model pruning during federated training. In our experiments in centralized and federated settings on the brain age prediction task (estimating a person's age from their brain MRI), we demonstrate that models can be pruned up to 95% sparsity without affecting performance even in challenging federated learning environments with highly heterogeneous data distributions. One surprising benefit of model pruning is improved model privacy. We demonstrate that models with high sparsity are less susceptible to membership inference attacks, a type of privacy attack.
Secure Federated Learning for Neuroimaging
May 11, 2022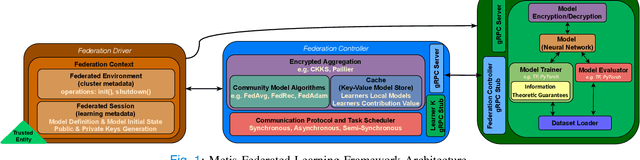
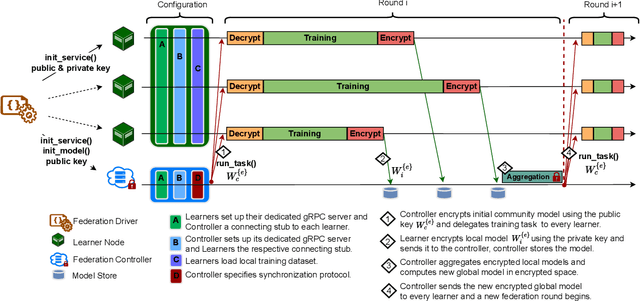
Abstract:The amount of biomedical data continues to grow rapidly. However, the ability to collect data from multiple sites for joint analysis remains challenging due to security, privacy, and regulatory concerns. We present a Secure Federated Learning architecture, MetisFL, which enables distributed training of neural networks over multiple data sources without sharing data. Each site trains the neural network over its private data for some time, then shares the neural network parameters (i.e., weights, gradients) with a Federation Controller, which in turn aggregates the local models, sends the resulting community model back to each site, and the process repeats. Our architecture provides strong security and privacy. First, sample data never leaves a site. Second, neural parameters are encrypted before transmission and the community model is computed under fully-homomorphic encryption. Finally, we use information-theoretic methods to limit information leakage from the neural model to prevent a curious site from performing membership attacks. We demonstrate this architecture in neuroimaging. Specifically, we investigate training neural models to classify Alzheimer's disease, and estimate Brain Age, from magnetic resonance imaging datasets distributed across multiple sites, including heterogeneous environments where sites have different amounts of data, statistical distributions, and computational capabilities.
Secure Neuroimaging Analysis using Federated Learning with Homomorphic Encryption
Aug 07, 2021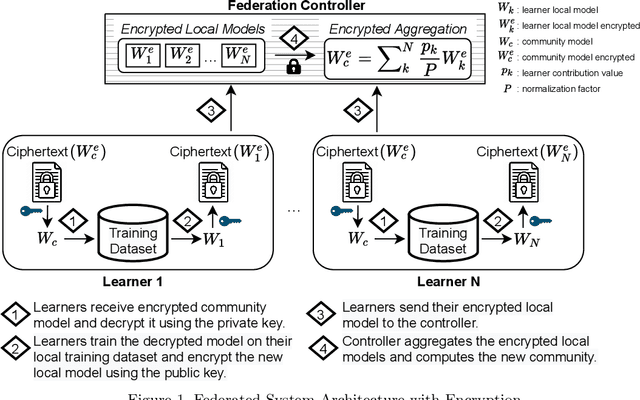
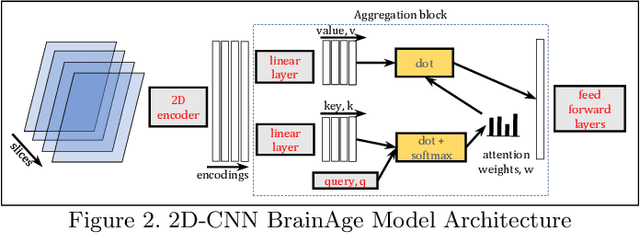
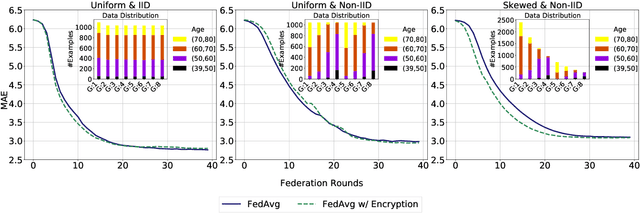
Abstract:Federated learning (FL) enables distributed computation of machine learning models over various disparate, remote data sources, without requiring to transfer any individual data to a centralized location. This results in an improved generalizability of models and efficient scaling of computation as more sources and larger datasets are added to the federation. Nevertheless, recent membership attacks show that private or sensitive personal data can sometimes be leaked or inferred when model parameters or summary statistics are shared with a central site, requiring improved security solutions. In this work, we propose a framework for secure FL using fully-homomorphic encryption (FHE). Specifically, we use the CKKS construction, an approximate, floating point compatible scheme that benefits from ciphertext packing and rescaling. In our evaluation on large-scale brain MRI datasets, we use our proposed secure FL framework to train a deep learning model to predict a person's age from distributed MRI scans, a common benchmarking task, and demonstrate that there is no degradation in the learning performance between the encrypted and non-encrypted federated models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge