Meng Hu
Streamlining Biomedical Research with Specialized LLMs
Apr 15, 2025Abstract:In this paper, we propose a novel system that integrates state-of-the-art, domain-specific large language models with advanced information retrieval techniques to deliver comprehensive and context-aware responses. Our approach facilitates seamless interaction among diverse components, enabling cross-validation of outputs to produce accurate, high-quality responses enriched with relevant data, images, tables, and other modalities. We demonstrate the system's capability to enhance response precision by leveraging a robust question-answering model, significantly improving the quality of dialogue generation. The system provides an accessible platform for real-time, high-fidelity interactions, allowing users to benefit from efficient human-computer interaction, precise retrieval, and simultaneous access to a wide range of literature and data. This dramatically improves the research efficiency of professionals in the biomedical and pharmaceutical domains and facilitates faster, more informed decision-making throughout the R\&D process. Furthermore, the system proposed in this paper is available at https://synapse-chat.patsnap.com.
Feature Statistics with Uncertainty Help Adversarial Robustness
Mar 26, 2025Abstract:Despite the remarkable success of deep neural networks (DNNs), the security threat of adversarial attacks poses a significant challenge to the reliability of DNNs. By introducing randomness into different parts of DNNs, stochastic methods can enable the model to learn some uncertainty, thereby improving model robustness efficiently. In this paper, we theoretically discover a universal phenomenon that adversarial attacks will shift the distributions of feature statistics. Motivated by this theoretical finding, we propose a robustness enhancement module called Feature Statistics with Uncertainty (FSU). It resamples channel-wise feature means and standard deviations of examples from multivariate Gaussian distributions, which helps to reconstruct the attacked examples and calibrate the shifted distributions. The calibration recovers some domain characteristics of the data for classification, thereby mitigating the influence of perturbations and weakening the ability of attacks to deceive models. The proposed FSU module has universal applicability in training, attacking, predicting and fine-tuning, demonstrating impressive robustness enhancement ability at trivial additional time cost. For example, against powerful optimization-based CW attacks, by incorporating FSU into attacking and predicting phases, it endows many collapsed state-of-the-art models with 50%-80% robust accuracy on CIFAR10, CIFAR100 and SVHN.
PharmaGPT: Domain-Specific Large Language Models for Bio-Pharmaceutical and Chemistry
Jul 03, 2024



Abstract:Large language models (LLMs) have revolutionized Natural Language Processing (NLP) by by minimizing the need for complex feature engineering. However, the application of LLMs in specialized domains like biopharmaceuticals and chemistry remains largely unexplored. These fields are characterized by intricate terminologies, specialized knowledge, and a high demand for precision areas where general purpose LLMs often fall short. In this study, we introduce PharmGPT, a suite of multilingual LLMs with 13 billion and 70 billion parameters, specifically trained on a comprehensive corpus of hundreds of billions of tokens tailored to the Bio-Pharmaceutical and Chemical sectors. Our evaluation shows that PharmGPT matches or surpasses existing general models on key benchmarks, such as NAPLEX, demonstrating its exceptional capability in domain-specific tasks. This advancement establishes a new benchmark for LLMs in the Bio-Pharmaceutical and Chemical fields, addressing the existing gap in specialized language modeling. Furthermore, this suggests a promising path for enhanced research and development in these specialized areas, paving the way for more precise and effective applications of NLP in specialized domains.
PharmGPT: Domain-Specific Large Language Models for Bio-Pharmaceutical and Chemistry
Jun 26, 2024



Abstract:Large language models (LLMs) have revolutionized Natural Language Processing (NLP) by by minimizing the need for complex feature engineering. However, the application of LLMs in specialized domains like biopharmaceuticals and chemistry remains largely unexplored. These fields are characterized by intricate terminologies, specialized knowledge, and a high demand for precision areas where general purpose LLMs often fall short. In this study, we introduce PharmGPT, a suite of multilingual LLMs with 13 billion and 70 billion parameters, specifically trained on a comprehensive corpus of hundreds of billions of tokens tailored to the Bio-Pharmaceutical and Chemical sectors. Our evaluation shows that PharmGPT matches or surpasses existing general models on key benchmarks, such as NAPLEX, demonstrating its exceptional capability in domain-specific tasks. This advancement establishes a new benchmark for LLMs in the Bio-Pharmaceutical and Chemical fields, addressing the existing gap in specialized language modeling. Furthermore, this suggests a promising path for enhanced research and development in these specialized areas, paving the way for more precise and effective applications of NLP in specialized domains.
PatentGPT: A Large Language Model for Intellectual Property
Apr 30, 2024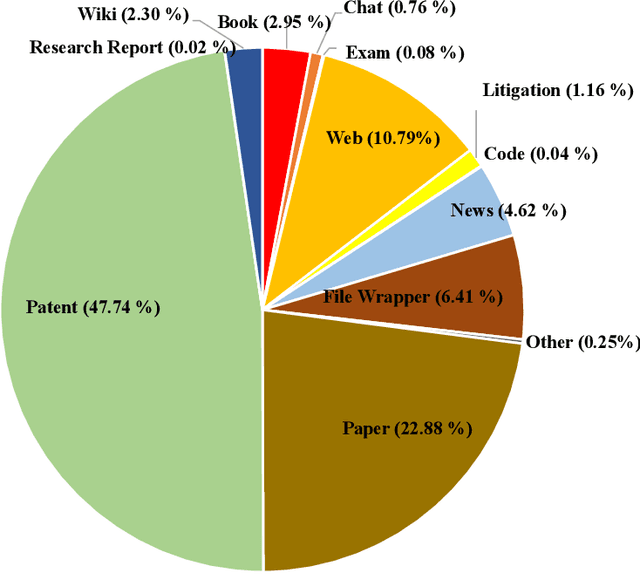

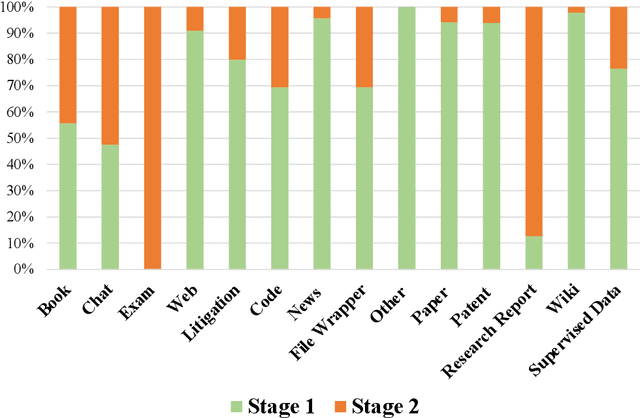
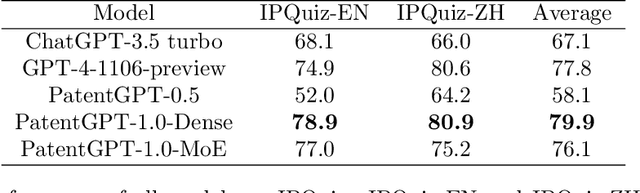
Abstract:In recent years, large language models have attracted significant attention due to their exceptional performance across a multitude of natural language process tasks, and have been widely applied in various fields. However, the application of large language models in the Intellectual Property (IP) space is challenging due to the strong need for specialized knowledge, privacy protection, processing of extremely long text in this field. In this technical report, we present for the first time a low-cost, standardized procedure for training IP-oriented LLMs, meeting the unique requirements of the IP domain. Using this standard process, we have trained the PatentGPT series models based on open-source pretrained models. By evaluating them on the open-source IP-oriented benchmark MOZIP, our domain-specific LLMs outperforms GPT-4, indicating the effectiveness of the proposed training procedure and the expertise of the PatentGPT models in the IP demain. What is impressive is that our model significantly outperformed GPT-4 on the 2019 China Patent Agent Qualification Examination by achieving a score of 65, reaching the level of human experts. Additionally, the PatentGPT model, which utilizes the SMoE architecture, achieves performance comparable to that of GPT-4 in the IP domain and demonstrates a better cost-performance ratio on long-text tasks, potentially serving as an alternative to GPT-4 within the IP domain.
Leveraging GPT-4 for Food Effect Summarization to Enhance Product-Specific Guidance Development via Iterative Prompting
Jun 28, 2023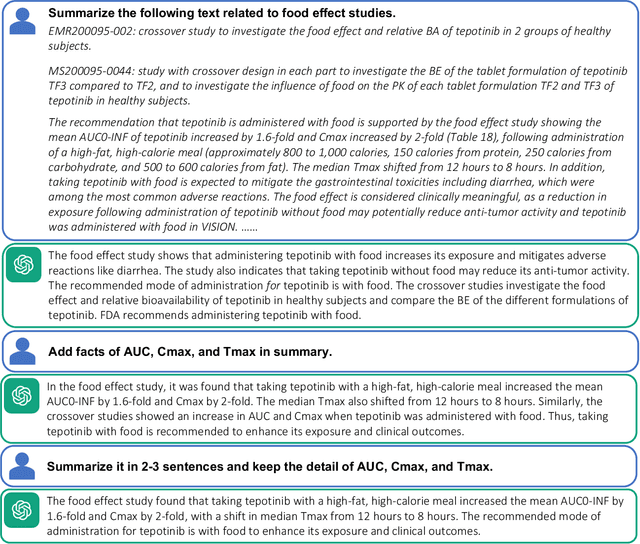
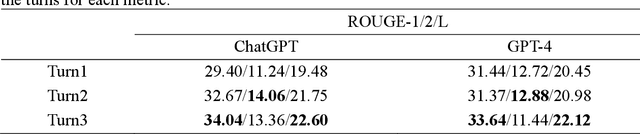

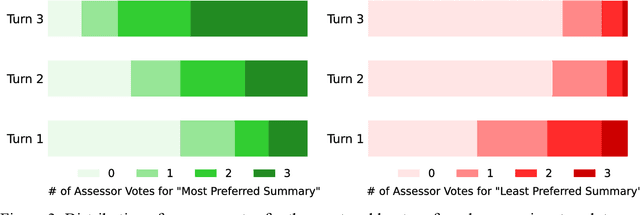
Abstract:Food effect summarization from New Drug Application (NDA) is an essential component of product-specific guidance (PSG) development and assessment. However, manual summarization of food effect from extensive drug application review documents is time-consuming, which arouses a need to develop automated methods. Recent advances in large language models (LLMs) such as ChatGPT and GPT-4, have demonstrated great potential in improving the effectiveness of automated text summarization, but its ability regarding the accuracy in summarizing food effect for PSG assessment remains unclear. In this study, we introduce a simple yet effective approach, iterative prompting, which allows one to interact with ChatGPT or GPT-4 more effectively and efficiently through multi-turn interaction. Specifically, we propose a three-turn iterative prompting approach to food effect summarization in which the keyword-focused and length-controlled prompts are respectively provided in consecutive turns to refine the quality of the generated summary. We conduct a series of extensive evaluations, ranging from automated metrics to FDA professionals and even evaluation by GPT-4, on 100 NDA review documents selected over the past five years. We observe that the summary quality is progressively improved throughout the process. Moreover, we find that GPT-4 performs better than ChatGPT, as evaluated by FDA professionals (43% vs. 12%) and GPT-4 (64% vs. 35%). Importantly, all the FDA professionals unanimously rated that 85% of the summaries generated by GPT-4 are factually consistent with the golden reference summary, a finding further supported by GPT-4 rating of 72% consistency. These results strongly suggest a great potential for GPT-4 to draft food effect summaries that could be reviewed by FDA professionals, thereby improving the efficiency of PSG assessment cycle and promoting the generic drug product development.
Fine-Tuning BERT for Automatic ADME Semantic Labeling in FDA Drug Labeling to Enhance Product-Specific Guidance Assessment
Jul 25, 2022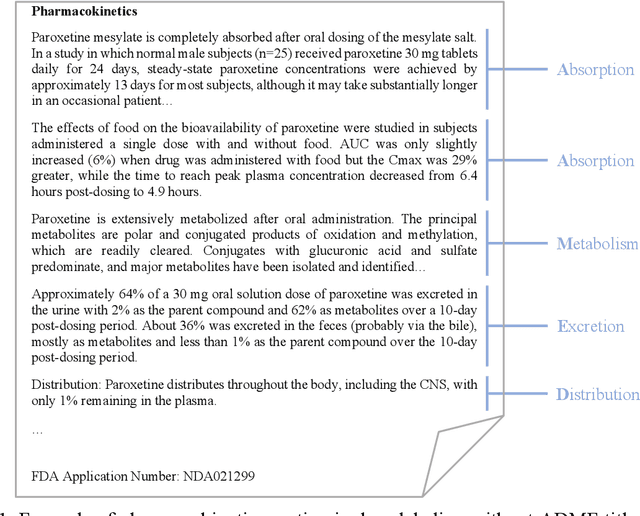

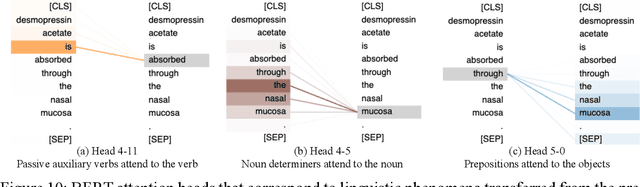
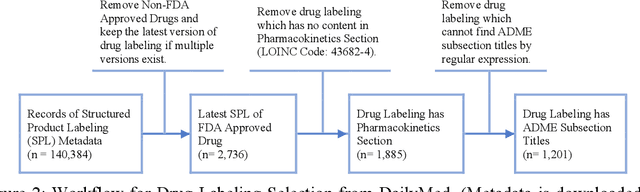
Abstract:Product-specific guidances (PSGs) recommended by the United States Food and Drug Administration (FDA) are instrumental to promote and guide generic drug product development. To assess a PSG, the FDA assessor needs to take extensive time and effort to manually retrieve supportive drug information of absorption, distribution, metabolism, and excretion (ADME) from the reference listed drug labeling. In this work, we leveraged the state-of-the-art pre-trained language models to automatically label the ADME paragraphs in the pharmacokinetics section from the FDA-approved drug labeling to facilitate PSG assessment. We applied a transfer learning approach by fine-tuning the pre-trained Bidirectional Encoder Representations from Transformers (BERT) model to develop a novel application of ADME semantic labeling, which can automatically retrieve ADME paragraphs from drug labeling instead of manual work. We demonstrated that fine-tuning the pre-trained BERT model can outperform the conventional machine learning techniques, achieving up to 11.6% absolute F1 improvement. To our knowledge, we were the first to successfully apply BERT to solve the ADME semantic labeling task. We further assessed the relative contribution of pre-training and fine-tuning to the overall performance of the BERT model in the ADME semantic labeling task using a series of analysis methods such as attention similarity and layer-based ablations. Our analysis revealed that the information learned via fine-tuning is focused on task-specific knowledge in the top layers of the BERT, whereas the benefit from the pre-trained BERT model is from the bottom layers.
Two-Stage Fine-Tuning: A Novel Strategy for Learning Class-Imbalanced Data
Jul 22, 2022
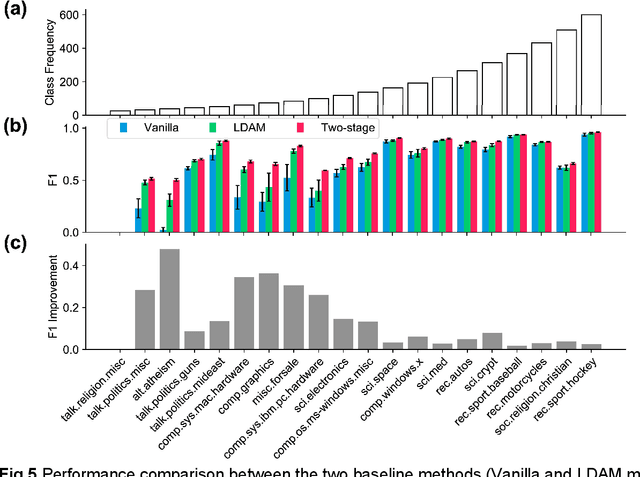
Abstract:Classification on long-tailed distributed data is a challenging problem, which suffers from serious class-imbalance and hence poor performance on tail classes with only a few samples. Owing to this paucity of samples, learning on the tail classes is especially challenging for the fine-tuning when transferring a pretrained model to a downstream task. In this work, we present a simple modification of standard fine-tuning to cope with these challenges. Specifically, we propose a two-stage fine-tuning: we first fine-tune the final layer of the pretrained model with class-balanced reweighting loss, and then we perform the standard fine-tuning. Our modification has several benefits: (1) it leverages pretrained representations by only fine-tuning a small portion of the model parameters while keeping the rest untouched; (2) it allows the model to learn an initial representation of the specific task; and importantly (3) it protects the learning of tail classes from being at a disadvantage during the model updating. We conduct extensive experiments on synthetic datasets of both two-class and multi-class tasks of text classification as well as a real-world application to ADME (i.e., absorption, distribution, metabolism, and excretion) semantic labeling. The experimental results show that the proposed two-stage fine-tuning outperforms both fine-tuning with conventional loss and fine-tuning with a reweighting loss on the above datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge