Mathieu Chevalley
In-silico biological discovery with large perturbation models
Mar 30, 2025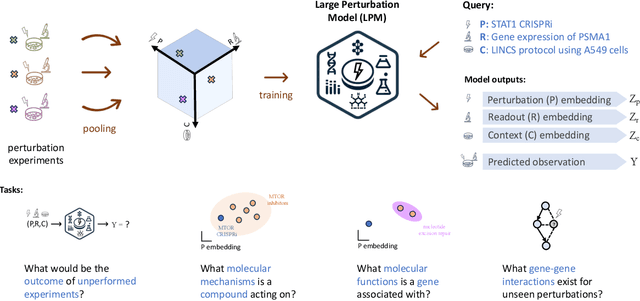
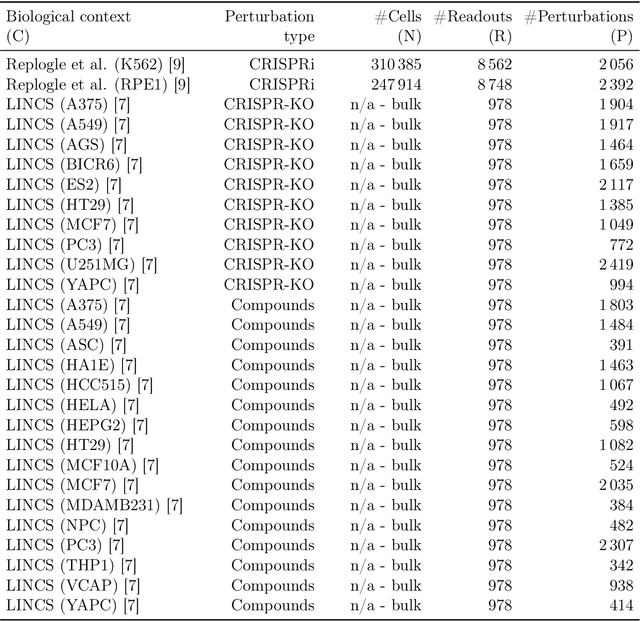
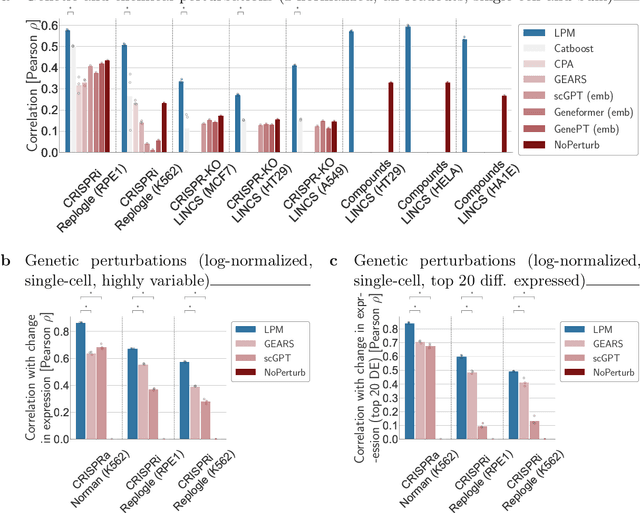
Abstract:Data generated in perturbation experiments link perturbations to the changes they elicit and therefore contain information relevant to numerous biological discovery tasks -- from understanding the relationships between biological entities to developing therapeutics. However, these data encompass diverse perturbations and readouts, and the complex dependence of experimental outcomes on their biological context makes it challenging to integrate insights across experiments. Here, we present the Large Perturbation Model (LPM), a deep-learning model that integrates multiple, heterogeneous perturbation experiments by representing perturbation, readout, and context as disentangled dimensions. LPM outperforms existing methods across multiple biological discovery tasks, including in predicting post-perturbation transcriptomes of unseen experiments, identifying shared molecular mechanisms of action between chemical and genetic perturbations, and facilitating the inference of gene-gene interaction networks.
Multi-megabase scale genome interpretation with genetic language models
Jan 13, 2025Abstract:Understanding how molecular changes caused by genetic variation drive disease risk is crucial for deciphering disease mechanisms. However, interpreting genome sequences is challenging because of the vast size of the human genome, and because its consequences manifest across a wide range of cells, tissues and scales -- spanning from molecular to whole organism level. Here, we present Phenformer, a multi-scale genetic language model that learns to generate mechanistic hypotheses as to how differences in genome sequence lead to disease-relevant changes in expression across cell types and tissues directly from DNA sequences of up to 88 million base pairs. Using whole genome sequencing data from more than 150 000 individuals, we show that Phenformer generates mechanistic hypotheses about disease-relevant cell and tissue types that match literature better than existing state-of-the-art methods, while using only sequence data. Furthermore, disease risk predictors enriched by Phenformer show improved prediction performance and generalisation to diverse populations. Accurate multi-megabase scale interpretation of whole genomes without additional experimental data enables both a deeper understanding of molecular mechanisms involved in disease and improved disease risk prediction at the level of individuals.
Efficient Differentiable Discovery of Causal Order
Oct 11, 2024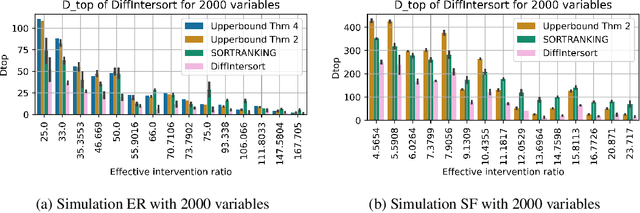
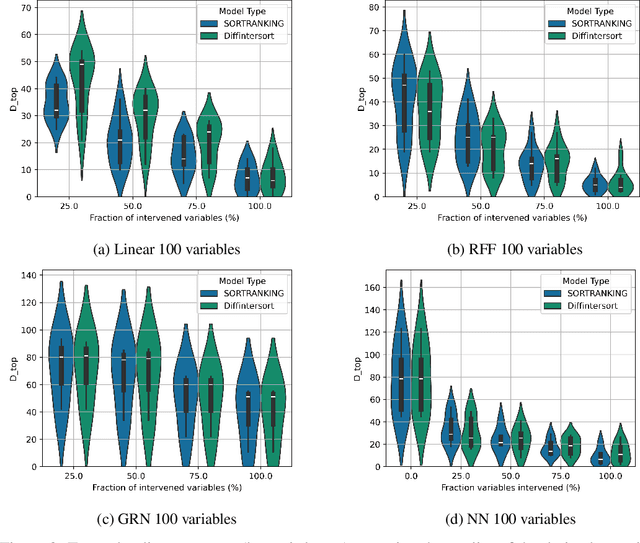
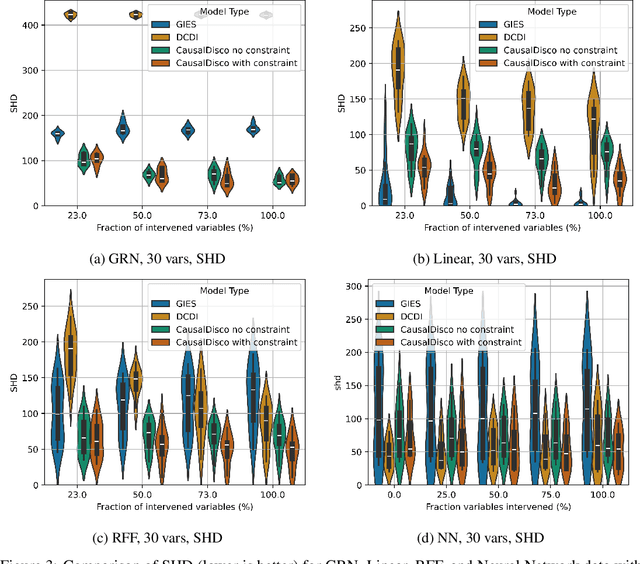
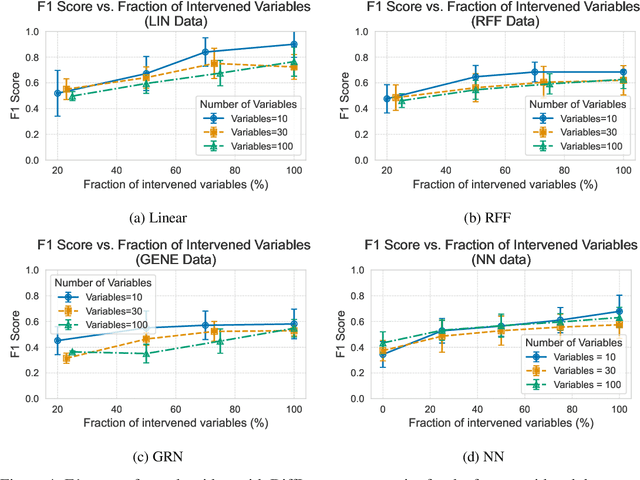
Abstract:In the algorithm Intersort, Chevalley et al. (2024) proposed a score-based method to discover the causal order of variables in a Directed Acyclic Graph (DAG) model, leveraging interventional data to outperform existing methods. However, as a score-based method over the permutahedron, Intersort is computationally expensive and non-differentiable, limiting its ability to be utilised in problems involving large-scale datasets, such as those in genomics and climate models, or to be integrated into end-to-end gradient-based learning frameworks. We address this limitation by reformulating Intersort using differentiable sorting and ranking techniques. Our approach enables scalable and differentiable optimization of causal orderings, allowing the continuous score function to be incorporated as a regularizer in downstream tasks. Empirical results demonstrate that causal discovery algorithms benefit significantly from regularizing on the causal order, underscoring the effectiveness of our method. Our work opens the door to efficiently incorporating regularization for causal order into the training of differentiable models and thereby addresses a long-standing limitation of purely associational supervised learning.
Deriving Causal Order from Single-Variable Interventions: Guarantees & Algorithm
May 28, 2024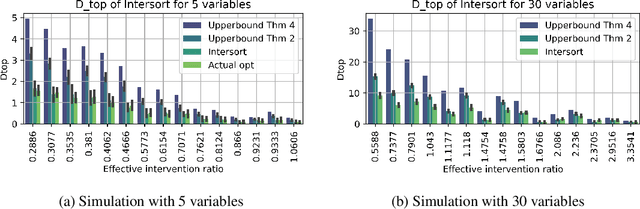
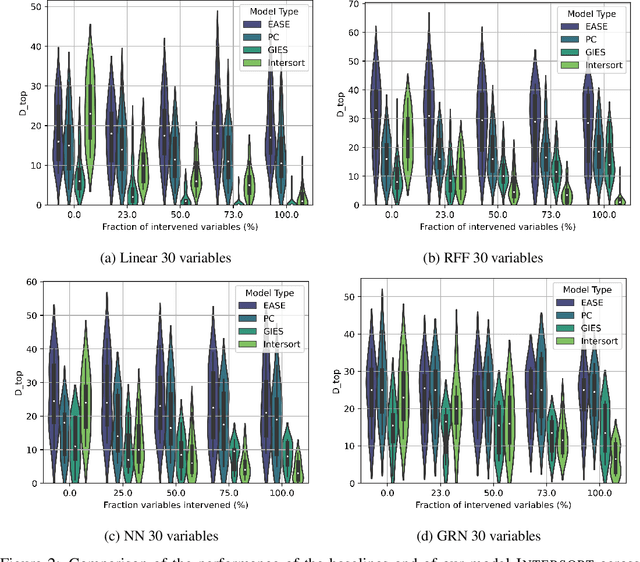
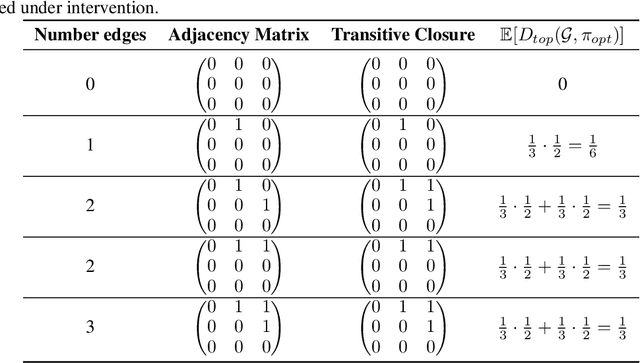
Abstract:Targeted and uniform interventions to a system are crucial for unveiling causal relationships. While several methods have been developed to leverage interventional data for causal structure learning, their practical application in real-world scenarios often remains challenging. Recent benchmark studies have highlighted these difficulties, even when large numbers of single-variable intervention samples are available. In this work, we demonstrate, both theoretically and empirically, that such datasets contain a wealth of causal information that can be effectively extracted under realistic assumptions about the data distribution. More specifically, we introduce the notion of interventional faithfulness, which relies on comparisons between the marginal distributions of each variable across observational and interventional settings, and we introduce a score on causal orders. Under this assumption, we are able to prove strong theoretical guarantees on the optimum of our score that also hold for large-scale settings. To empirically verify our theory, we introduce Intersort, an algorithm designed to infer the causal order from datasets containing large numbers of single-variable interventions by approximately optimizing our score. Intersort outperforms baselines (GIES, PC and EASE) on almost all simulated data settings replicating common benchmarks in the field. Our proposed novel approach to modeling interventional datasets thus offers a promising avenue for advancing causal inference, highlighting significant potential for further enhancements under realistic assumptions.
The CausalBench challenge: A machine learning contest for gene network inference from single-cell perturbation data
Aug 29, 2023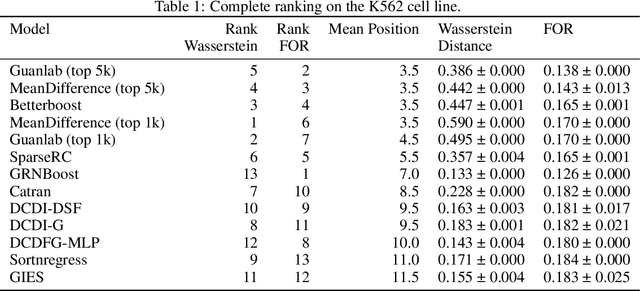
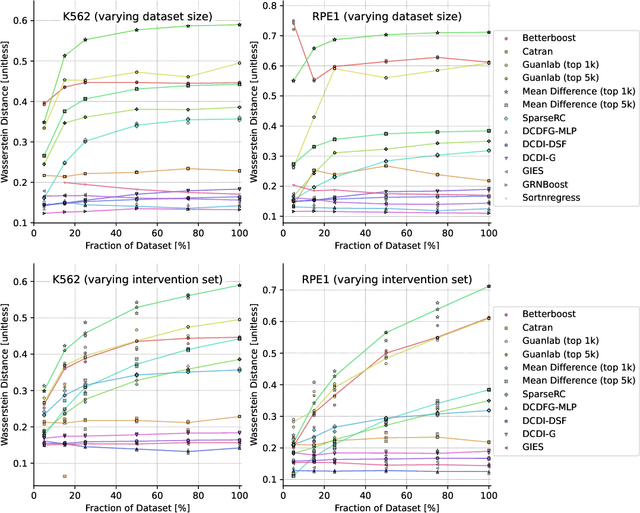
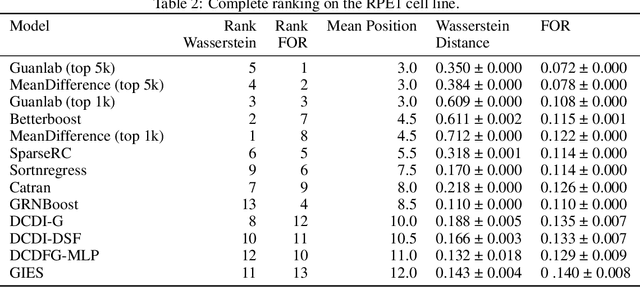
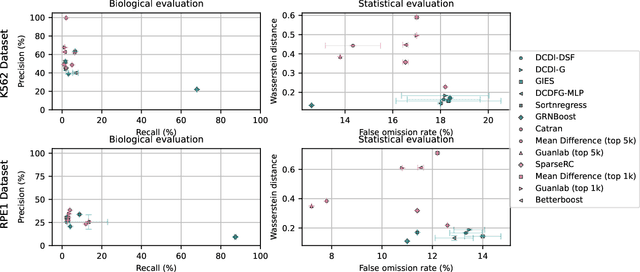
Abstract:In drug discovery, mapping interactions between genes within cellular systems is a crucial early step. This helps formulate hypotheses regarding molecular mechanisms that could potentially be targeted by future medicines. The CausalBench Challenge was an initiative to invite the machine learning community to advance the state of the art in constructing gene-gene interaction networks. These networks, derived from large-scale, real-world datasets of single cells under various perturbations, are crucial for understanding the causal mechanisms underlying disease biology. Using the framework provided by the CausalBench benchmark, participants were tasked with enhancing the capacity of the state of the art methods to leverage large-scale genetic perturbation data. This report provides an analysis and summary of the methods submitted during the challenge to give a partial image of the state of the art at the time of the challenge. The winning solutions significantly improved performance compared to previous baselines, establishing a new state of the art for this critical task in biology and medicine.
CausalBench: A Large-scale Benchmark for Network Inference from Single-cell Perturbation Data
Oct 31, 2022Abstract:Mapping biological mechanisms in cellular systems is a fundamental step in early-stage drug discovery that serves to generate hypotheses on what disease-relevant molecular targets may effectively be modulated by pharmacological interventions. With the advent of high-throughput methods for measuring single-cell gene expression under genetic perturbations, we now have effective means for generating evidence for causal gene-gene interactions at scale. However, inferring graphical networks of the size typically encountered in real-world gene-gene interaction networks is difficult in terms of both achieving and evaluating faithfulness to the true underlying causal graph. Moreover, standardised benchmarks for comparing methods for causal discovery in perturbational single-cell data do not yet exist. Here, we introduce CausalBench - a comprehensive benchmark suite for evaluating network inference methods on large-scale perturbational single-cell gene expression data. CausalBench introduces several biologically meaningful performance metrics and operates on two large, curated and openly available benchmark data sets for evaluating methods on the inference of gene regulatory networks from single-cell data generated under perturbations. With real-world datasets consisting of over \numprint{200000} training samples under interventions, CausalBench could potentially help facilitate advances in causal network inference by providing what is - to the best of our knowledge - the largest openly available test bed for causal discovery from real-world perturbation data to date.
Invariant Causal Mechanisms through Distribution Matching
Jun 23, 2022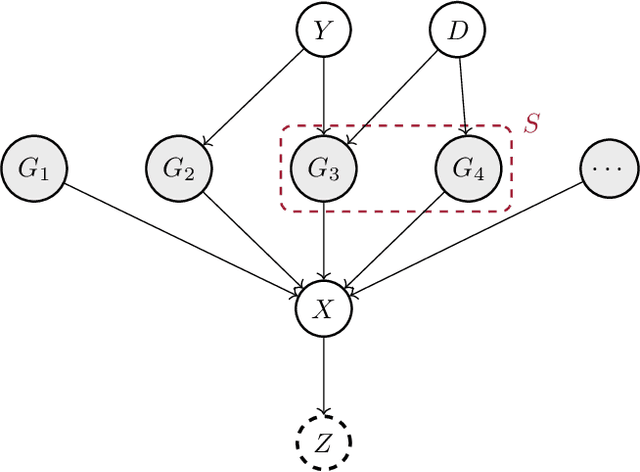
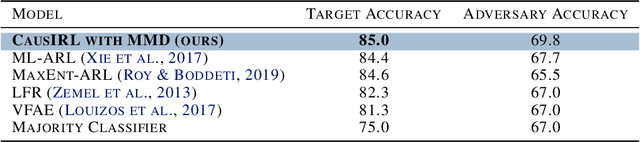
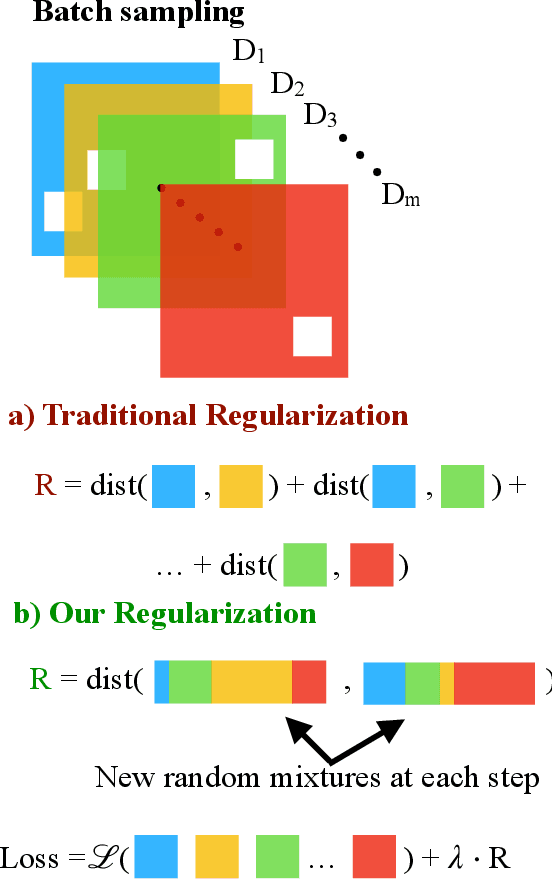
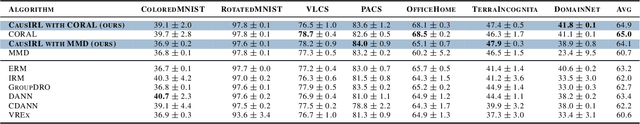
Abstract:Learning representations that capture the underlying data generating process is a key problem for data efficient and robust use of neural networks. One key property for robustness which the learned representation should capture and which recently received a lot of attention is described by the notion of invariance. In this work we provide a causal perspective and new algorithm for learning invariant representations. Empirically we show that this algorithm works well on a diverse set of tasks and in particular we observe state-of-the-art performance on domain generalization, where we are able to significantly boost the score of existing models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge