Dariusz Brzezinski
Among Them: A game-based framework for assessing persuasion capabilities of LLMs
Feb 27, 2025Abstract:The proliferation of large language models (LLMs) and autonomous AI agents has raised concerns about their potential for automated persuasion and social influence. While existing research has explored isolated instances of LLM-based manipulation, systematic evaluations of persuasion capabilities across different models remain limited. In this paper, we present an Among Us-inspired game framework for assessing LLM deception skills in a controlled environment. The proposed framework makes it possible to compare LLM models by game statistics, as well as quantify in-game manipulation according to 25 persuasion strategies from social psychology and rhetoric. Experiments between 8 popular language models of different types and sizes demonstrate that all tested models exhibit persuasive capabilities, successfully employing 22 of the 25 anticipated techniques. We also find that larger models do not provide any persuasion advantage over smaller models and that longer model outputs are negatively correlated with the number of games won. Our study provides insights into the deception capabilities of LLMs, as well as tools and data for fostering future research on the topic.
Random Similarity Isolation Forests
Feb 26, 2025Abstract:With predictive models becoming prevalent, companies are expanding the types of data they gather. As a result, the collected datasets consist not only of simple numerical features but also more complex objects such as time series, images, or graphs. Such multi-modal data have the potential to improve performance in predictive tasks like outlier detection, where the goal is to identify objects deviating from the main data distribution. However, current outlier detection algorithms are dedicated to individual types of data. Consequently, working with mixed types of data requires either fusing multiple data-specific models or transforming all of the representations into a single format, both of which can hinder predictive performance. In this paper, we propose a multi-modal outlier detection algorithm called Random Similarity Isolation Forest. Our method combines the notions of isolation and similarity-based projection to handle datasets with mixtures of features of arbitrary data types. Experiments performed on 47 benchmark datasets demonstrate that Random Similarity Isolation Forest outperforms five state-of-the-art competitors. Our study shows that the use of multiple modalities can indeed improve the detection of anomalies and highlights the need for new outlier detection benchmarks tailored for multi-modal algorithms.
Properties of fairness measures in the context of varying class imbalance and protected group ratios
Nov 13, 2024
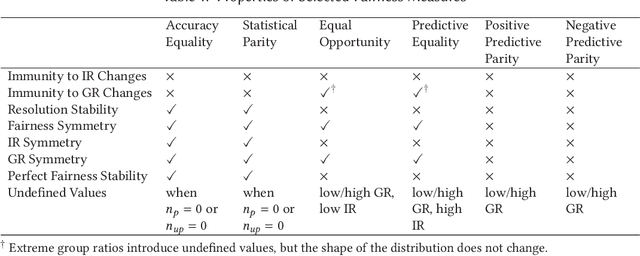
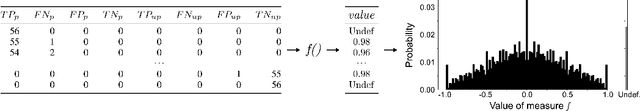
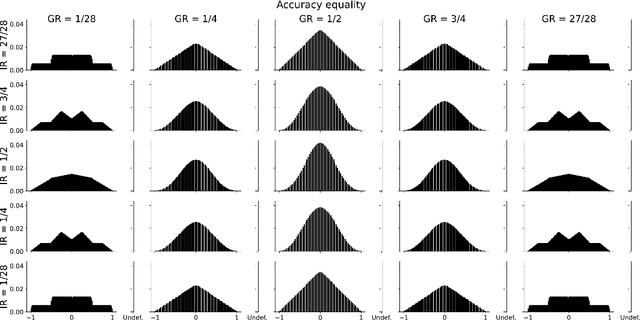
Abstract:Society is increasingly relying on predictive models in fields like criminal justice, credit risk management, or hiring. To prevent such automated systems from discriminating against people belonging to certain groups, fairness measures have become a crucial component in socially relevant applications of machine learning. However, existing fairness measures have been designed to assess the bias between predictions for protected groups without considering the imbalance in the classes of the target variable. Current research on the potential effect of class imbalance on fairness focuses on practical applications rather than dataset-independent measure properties. In this paper, we study the general properties of fairness measures for changing class and protected group proportions. For this purpose, we analyze the probability mass functions of six of the most popular group fairness measures. We also measure how the probability of achieving perfect fairness changes for varying class imbalance ratios. Moreover, we relate the dataset-independent properties of fairness measures described in this paper to classifier fairness in real-life tasks. Our results show that measures such as Equal Opportunity and Positive Predictive Parity are more sensitive to changes in class imbalance than Accuracy Equality. These findings can help guide researchers and practitioners in choosing the most appropriate fairness measures for their classification problems.
ART: Actually Robust Training
Aug 29, 2024Abstract:Current interest in deep learning captures the attention of many programmers and researchers. Unfortunately, the lack of a unified schema for developing deep learning models results in methodological inconsistencies, unclear documentation, and problems with reproducibility. Some guidelines have been proposed, yet currently, they lack practical implementations. Furthermore, neural network training often takes on the form of trial and error, lacking a structured and thoughtful process. To alleviate these issues, in this paper, we introduce Art, a Python library designed to help automatically impose rules and standards while developing deep learning pipelines. Art divides model development into a series of smaller steps of increasing complexity, each concluded with a validation check improving the interpretability and robustness of the process. The current version of Art comes equipped with nine predefined steps inspired by Andrej Karpathy's Recipe for Training Neural Networks, a visualization dashboard, and integration with loggers such as Neptune. The code related to this paper is available at: https://github.com/SebChw/Actually-Robust-Training.
The CausalBench challenge: A machine learning contest for gene network inference from single-cell perturbation data
Aug 29, 2023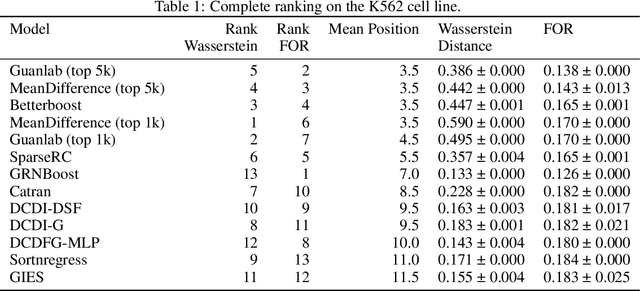
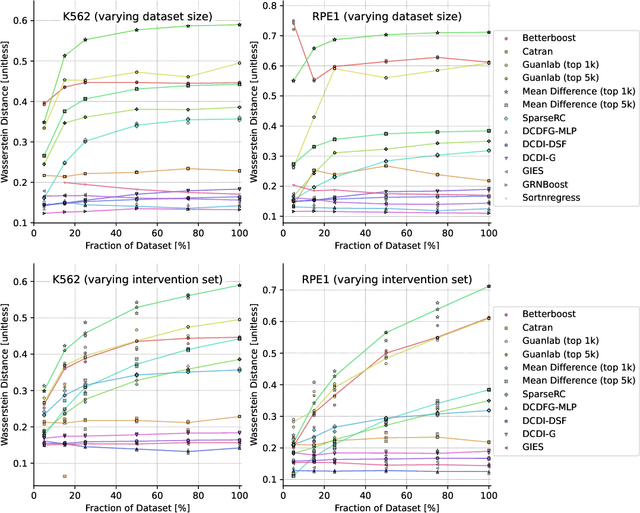
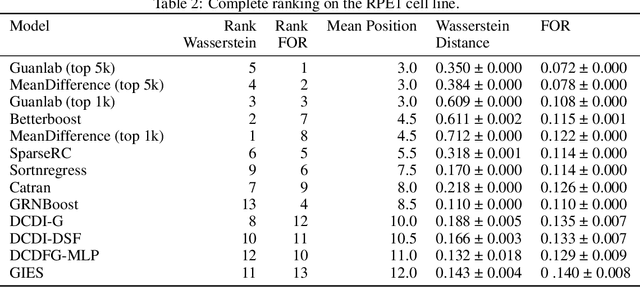
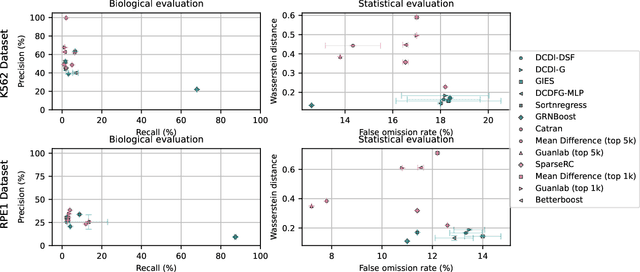
Abstract:In drug discovery, mapping interactions between genes within cellular systems is a crucial early step. This helps formulate hypotheses regarding molecular mechanisms that could potentially be targeted by future medicines. The CausalBench Challenge was an initiative to invite the machine learning community to advance the state of the art in constructing gene-gene interaction networks. These networks, derived from large-scale, real-world datasets of single cells under various perturbations, are crucial for understanding the causal mechanisms underlying disease biology. Using the framework provided by the CausalBench benchmark, participants were tasked with enhancing the capacity of the state of the art methods to leverage large-scale genetic perturbation data. This report provides an analysis and summary of the methods submitted during the challenge to give a partial image of the state of the art at the time of the challenge. The winning solutions significantly improved performance compared to previous baselines, establishing a new state of the art for this critical task in biology and medicine.
Towards Explainable TOPSIS: Visual Insights into the Effects of Weights and Aggregations on Rankings
Jun 13, 2023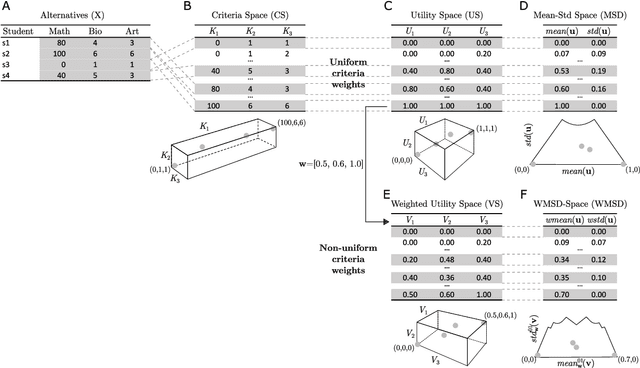
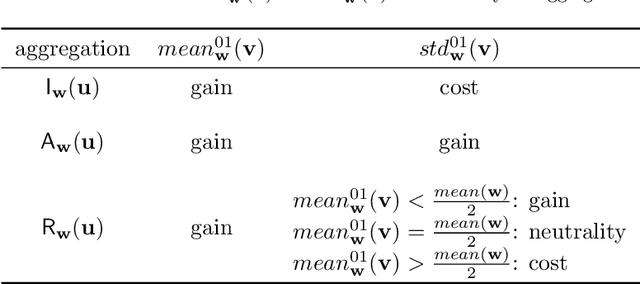
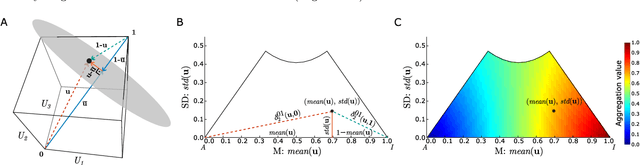
Abstract:Multi-Criteria Decision Analysis (MCDA) is extensively used across diverse industries to assess and rank alternatives. Among numerous MCDA methods developed to solve real-world ranking problems, TOPSIS remains one of the most popular choices in many application areas. TOPSIS calculates distances between the considered alternatives and two predefined ones, namely the ideal and the anti-ideal, and creates a ranking of the alternatives according to a chosen aggregation of these distances. However, the interpretation of the inner workings of TOPSIS is difficult, especially when the number of criteria is large. To this end, recent research has shown that TOPSIS aggregations can be expressed using the means (M) and standard deviations (SD) of alternatives, creating MSD-space, a tool for visualizing and explaining aggregations. Even though MSD-space is highly useful, it assumes equally important criteria, making it less applicable to real-world ranking problems. In this paper, we generalize the concept of MSD-space to weighted criteria by introducing the concept of WMSD-space defined by what is referred to as weight-scaled means and standard deviations. We demonstrate that TOPSIS and similar distance-based aggregation methods can be successfully illustrated in a plane and interpreted even when the criteria are weighted, regardless of their number. The proposed WMSD-space offers a practical method for explaining TOPSIS rankings in real-world decision problems.
confidence-planner: Easy-to-Use Prediction Confidence Estimation and Sample Size Planning
Jan 12, 2023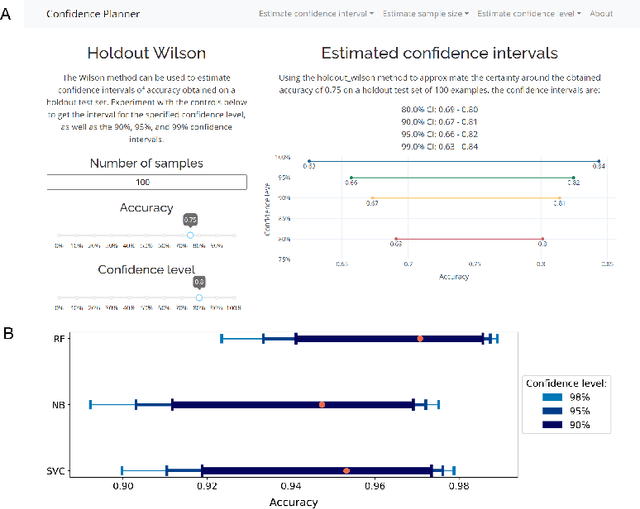
Abstract:Machine learning applications, especially in the fields of me\-di\-cine and social sciences, are slowly being subjected to increasing scrutiny. Similarly to sample size planning performed in clinical and social studies, lawmakers and funding agencies may expect statistical uncertainty estimations in machine learning applications that impact society. In this paper, we present an easy-to-use python package and web application for estimating prediction confidence intervals. The package offers eight different procedures to determine and justify the sample size and confidence of predictions from holdout, bootstrap, cross-validation, and progressive validation experiments. Since the package builds directly on established data analysis libraries, it seamlessly integrates into preprocessing and exploratory data analysis steps. Code related to this paper is available at: https://github.com/dabrze/confidence-planner.
Random Similarity Forests
Apr 11, 2022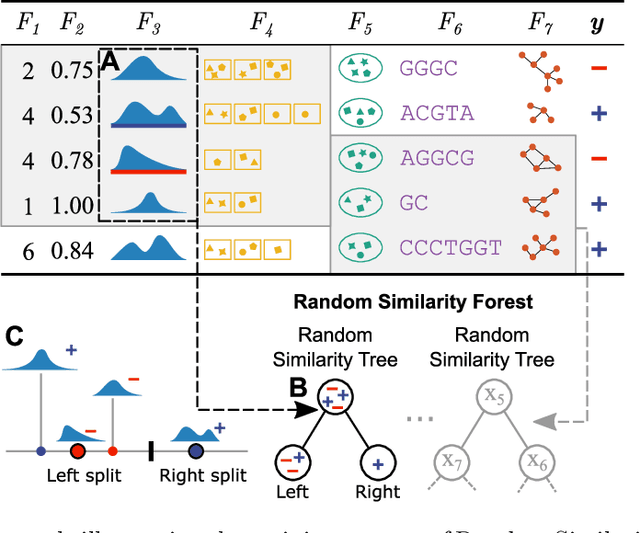
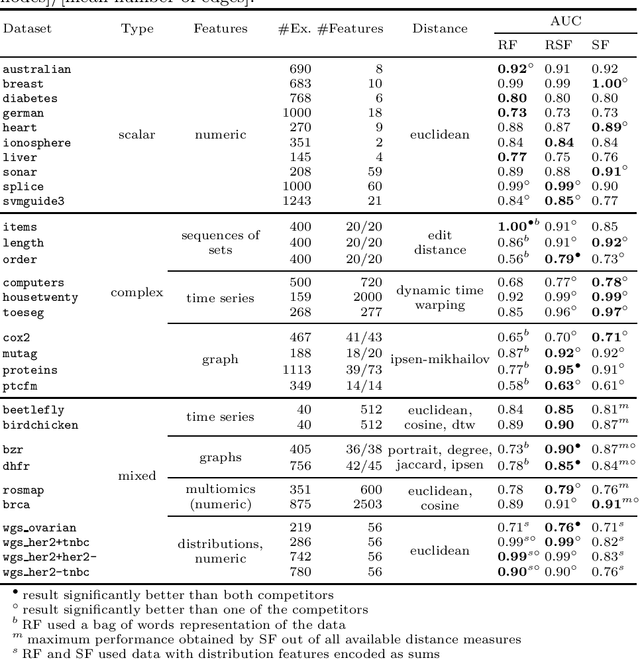
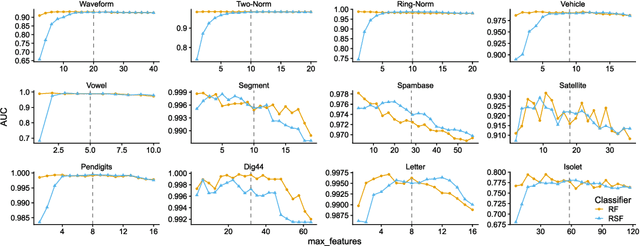
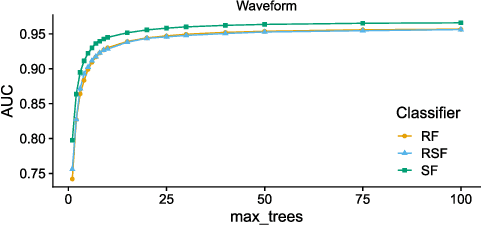
Abstract:The wealth of data being gathered about humans and their surroundings drives new machine learning applications in various fields. Consequently, more and more often, classifiers are trained using not only numerical data but also complex data objects. For example, multi-omics analyses attempt to combine numerical descriptions with distributions, time series data, discrete sequences, and graphs. Such integration of data from different domains requires either omitting some of the data, creating separate models for different formats, or simplifying some of the data to adhere to a shared scale and format, all of which can hinder predictive performance. In this paper, we propose a classification method capable of handling datasets with features of arbitrary data types while retaining each feature's characteristic. The proposed algorithm, called Random Similarity Forest, uses multiple domain-specific distance measures to combine the predictive performance of Random Forests with the flexibility of Similarity Forests. We show that Random Similarity Forests are on par with Random Forests on numerical data and outperform them on datasets from complex or mixed data domains. Our results highlight the applicability of Random Similarity Forests to noisy, multi-source datasets that are becoming ubiquitous in high-impact life science projects.
Fibonacci and k-Subsecting Recursive Feature Elimination
Jul 29, 2020



Abstract:Feature selection is a data mining task with the potential of speeding up classification algorithms, enhancing model comprehensibility, and improving learning accuracy. However, finding a subset of features that is optimal in terms of predictive accuracy is usually computationally intractable. Out of several heuristic approaches to dealing with this problem, the Recursive Feature Elimination (RFE) algorithm has received considerable interest from data mining practitioners. In this paper, we propose two novel algorithms inspired by RFE, called Fibonacci- and k-Subsecting Recursive Feature Elimination, which remove features in logarithmic steps, probing the wrapped classifier more densely for the more promising feature subsets. The proposed algorithms are experimentally compared against RFE on 28 highly multidimensional datasets and evaluated in a practical case study involving 3D electron density maps from the Protein Data Bank. The results show that Fibonacci and k-Subsecting Recursive Feature Elimination are capable of selecting a smaller subset of features much faster than standard RFE, while achieving comparable predictive performance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge