Andreas Georgiou
In-silico biological discovery with large perturbation models
Mar 30, 2025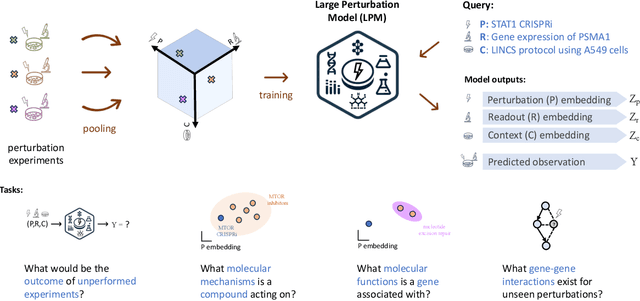
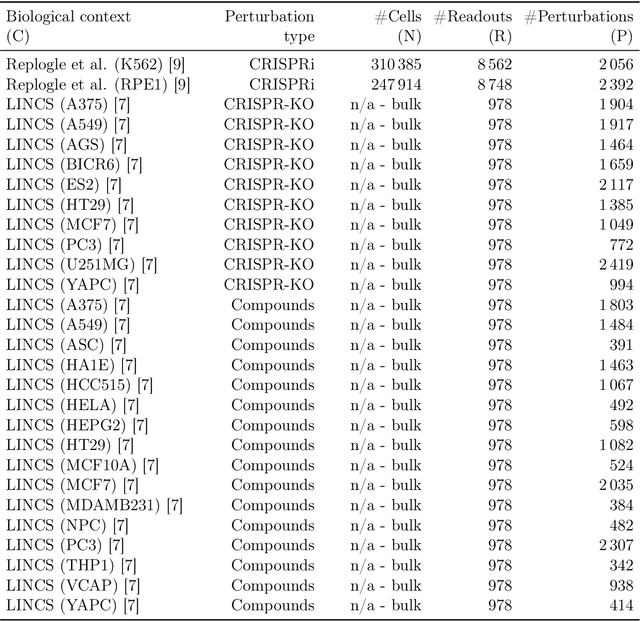
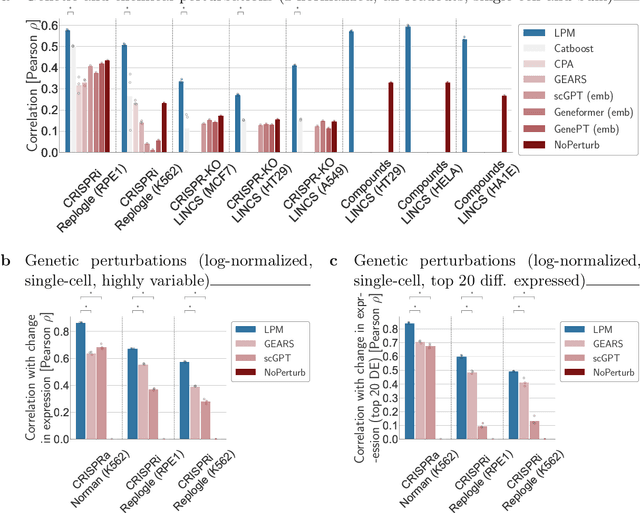
Abstract:Data generated in perturbation experiments link perturbations to the changes they elicit and therefore contain information relevant to numerous biological discovery tasks -- from understanding the relationships between biological entities to developing therapeutics. However, these data encompass diverse perturbations and readouts, and the complex dependence of experimental outcomes on their biological context makes it challenging to integrate insights across experiments. Here, we present the Large Perturbation Model (LPM), a deep-learning model that integrates multiple, heterogeneous perturbation experiments by representing perturbation, readout, and context as disentangled dimensions. LPM outperforms existing methods across multiple biological discovery tasks, including in predicting post-perturbation transcriptomes of unseen experiments, identifying shared molecular mechanisms of action between chemical and genetic perturbations, and facilitating the inference of gene-gene interaction networks.
Multi-megabase scale genome interpretation with genetic language models
Jan 13, 2025Abstract:Understanding how molecular changes caused by genetic variation drive disease risk is crucial for deciphering disease mechanisms. However, interpreting genome sequences is challenging because of the vast size of the human genome, and because its consequences manifest across a wide range of cells, tissues and scales -- spanning from molecular to whole organism level. Here, we present Phenformer, a multi-scale genetic language model that learns to generate mechanistic hypotheses as to how differences in genome sequence lead to disease-relevant changes in expression across cell types and tissues directly from DNA sequences of up to 88 million base pairs. Using whole genome sequencing data from more than 150 000 individuals, we show that Phenformer generates mechanistic hypotheses about disease-relevant cell and tissue types that match literature better than existing state-of-the-art methods, while using only sequence data. Furthermore, disease risk predictors enriched by Phenformer show improved prediction performance and generalisation to diverse populations. Accurate multi-megabase scale interpretation of whole genomes without additional experimental data enables both a deeper understanding of molecular mechanisms involved in disease and improved disease risk prediction at the level of individuals.
Enhanced Frame and Event-Based Simulator and Event-Based Video Interpolation Network
Dec 17, 2021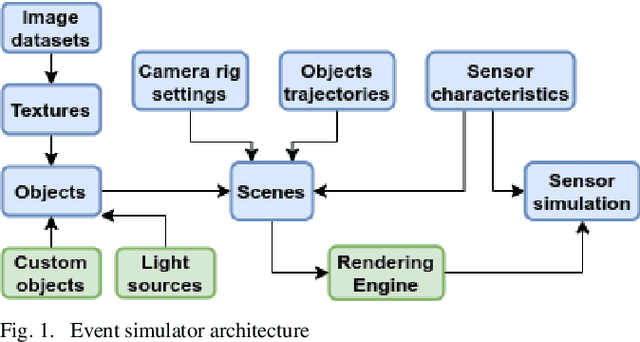
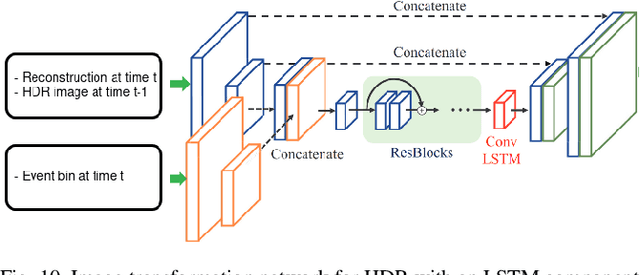
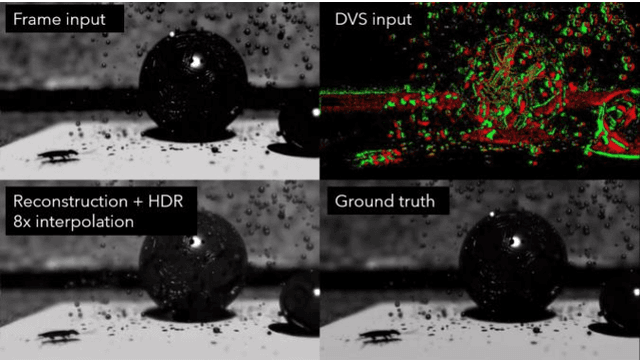

Abstract:Fast neuromorphic event-based vision sensors (Dynamic Vision Sensor, DVS) can be combined with slower conventional frame-based sensors to enable higher-quality inter-frame interpolation than traditional methods relying on fixed motion approximations using e.g. optical flow. In this work we present a new, advanced event simulator that can produce realistic scenes recorded by a camera rig with an arbitrary number of sensors located at fixed offsets. It includes a new configurable frame-based image sensor model with realistic image quality reduction effects, and an extended DVS model with more accurate characteristics. We use our simulator to train a novel reconstruction model designed for end-to-end reconstruction of high-fps video. Unlike previously published methods, our method does not require the frame and DVS cameras to have the same optics, positions, or camera resolutions. It is also not limited to objects a fixed distance from the sensor. We show that data generated by our simulator can be used to train our new model, leading to reconstructed images on public datasets of equivalent or better quality than the state of the art. We also show our sensor generalizing to data recorded by real sensors.
Deep Multiple Instance Learning for Taxonomic Classification of Metagenomic read sets
Sep 28, 2019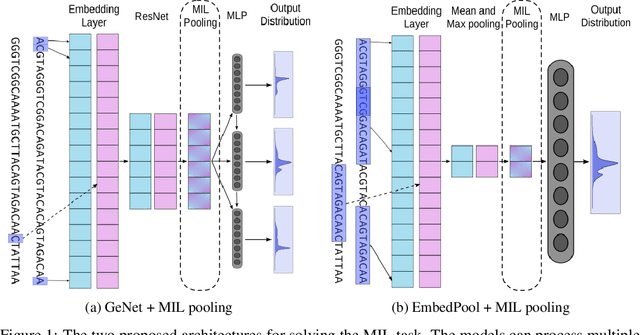
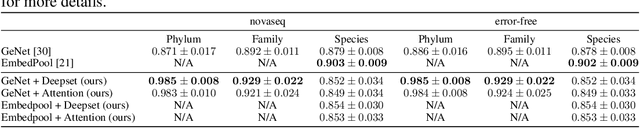
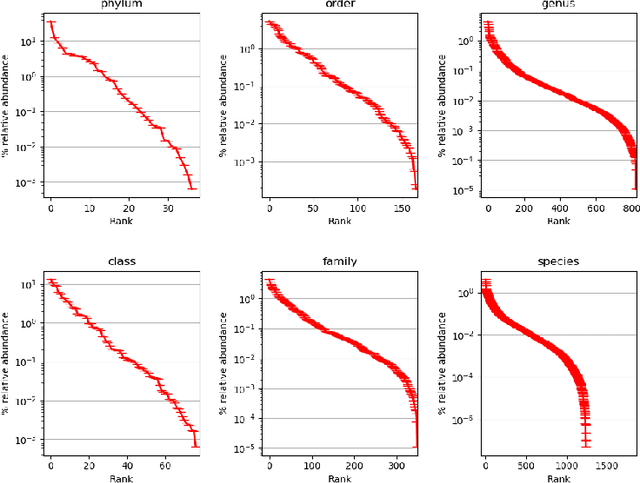
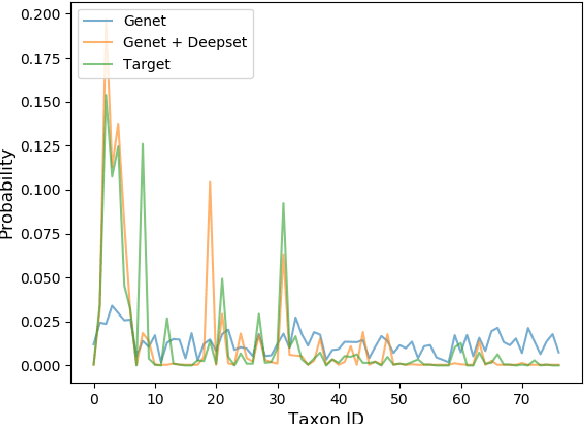
Abstract:Metagenomic studies have increasingly utilized sequencing technologies in order to analyze DNA fragments found in environmental samples. It can provide useful insights for studying the interactions between hosts and microbes, infectious disease proliferation, and novel species discovery. One important step in this analysis is the taxonomic classification of those DNA fragments. Of particular interest is the determination of the distribution of the taxa of microbes in metagenomic samples. Recent attempts using deep learning focus on architectures that classify single DNA reads independently from each other. In this work, we attempt to solve the task of directly predicting the distribution over the taxa of whole metagenomic read sets. We formulate this task as a Multiple Instance Learning (MIL) problem. We extend architectures used in single-read taxonomic classification with two different types of permutation-invariant MIL pooling layers: a) deepsets and b) attention-based pooling. We illustrate that our architecture can exploit the co-occurrence of species in metagenomic read sets and outperforms the single-read architectures in predicting the distribution over the taxa at higher taxonomic ranks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge