Gunnar Rätsch
Data-Driven Discovery of Feature Groups in Clinical Time Series
Nov 11, 2025Abstract:Clinical time series data are critical for patient monitoring and predictive modeling. These time series are typically multivariate and often comprise hundreds of heterogeneous features from different data sources. The grouping of features based on similarity and relevance to the prediction task has been shown to enhance the performance of deep learning architectures. However, defining these groups a priori using only semantic knowledge is challenging, even for domain experts. To address this, we propose a novel method that learns feature groups by clustering weights of feature-wise embedding layers. This approach seamlessly integrates into standard supervised training and discovers the groups that directly improve downstream performance on clinically relevant tasks. We demonstrate that our method outperforms static clustering approaches on synthetic data and achieves performance comparable to expert-defined groups on real-world medical data. Moreover, the learned feature groups are clinically interpretable, enabling data-driven discovery of task-relevant relationships between variables.
Domain Generalization and Adaptation in Intensive Care with Anchor Regression
Jul 29, 2025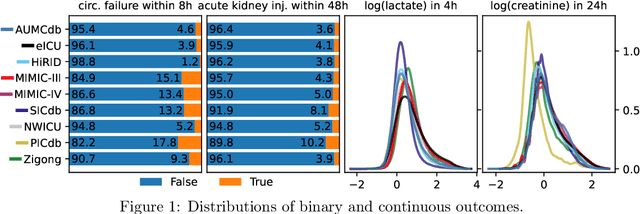

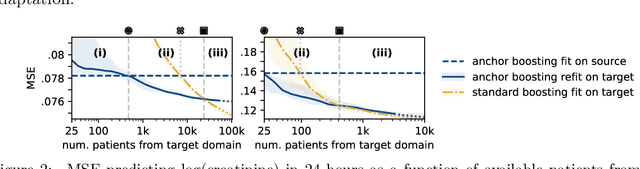
Abstract:The performance of predictive models in clinical settings often degrades when deployed in new hospitals due to distribution shifts. This paper presents a large-scale study of causality-inspired domain generalization on heterogeneous multi-center intensive care unit (ICU) data. We apply anchor regression and introduce anchor boosting, a novel, tree-based nonlinear extension, to a large dataset comprising 400,000 patients from nine distinct ICU databases. The anchor regularization consistently improves out-of-distribution performance, particularly for the most dissimilar target domains. The methods appear robust to violations of theoretical assumptions, such as anchor exogeneity. Furthermore, we propose a novel conceptual framework to quantify the utility of large external data datasets. By evaluating performance as a function of available target-domain data, we identify three regimes: (i) a domain generalization regime, where only the external model should be used, (ii) a domain adaptation regime, where refitting the external model is optimal, and (iii) a data-rich regime, where external data provides no additional value.
Revisiting Automatic Data Curation for Vision Foundation Models in Digital Pathology
Mar 24, 2025Abstract:Vision foundation models (FMs) are accelerating the development of digital pathology algorithms and transforming biomedical research. These models learn, in a self-supervised manner, to represent histological features in highly heterogeneous tiles extracted from whole-slide images (WSIs) of real-world patient samples. The performance of these FMs is significantly influenced by the size, diversity, and balance of the pre-training data. However, data selection has been primarily guided by expert knowledge at the WSI level, focusing on factors such as disease classification and tissue types, while largely overlooking the granular details available at the tile level. In this paper, we investigate the potential of unsupervised automatic data curation at the tile-level, taking into account 350 million tiles. Specifically, we apply hierarchical clustering trees to pre-extracted tile embeddings, allowing us to sample balanced datasets uniformly across the embedding space of the pretrained FM. We further identify these datasets are subject to a trade-off between size and balance, potentially compromising the quality of representations learned by FMs, and propose tailored batch sampling strategies to mitigate this effect. We demonstrate the effectiveness of our method through improved performance on a diverse range of clinically relevant downstream tasks.
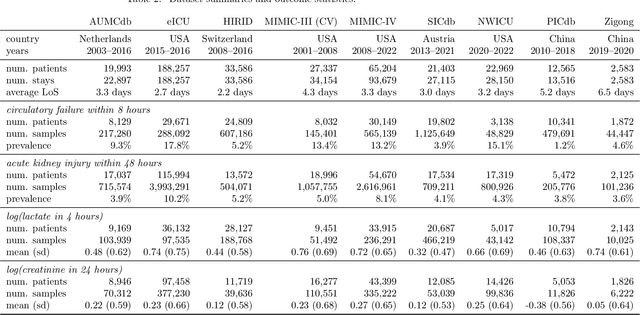
Towards Foundation Models for Critical Care Time Series
Nov 25, 2024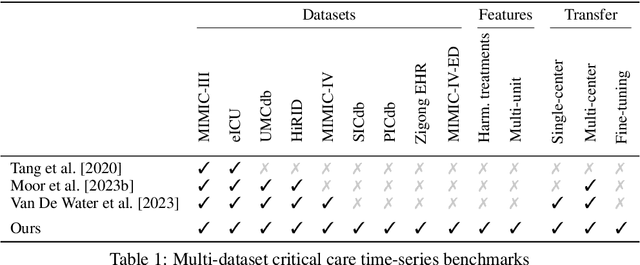
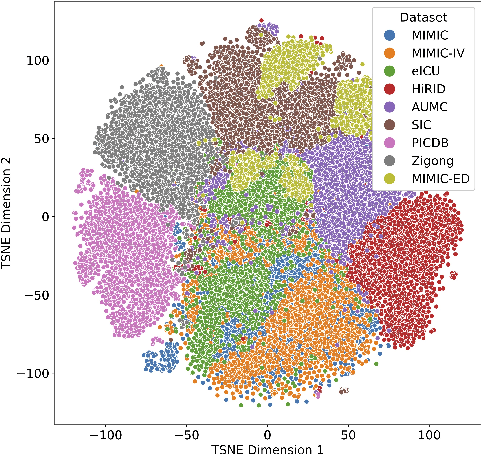
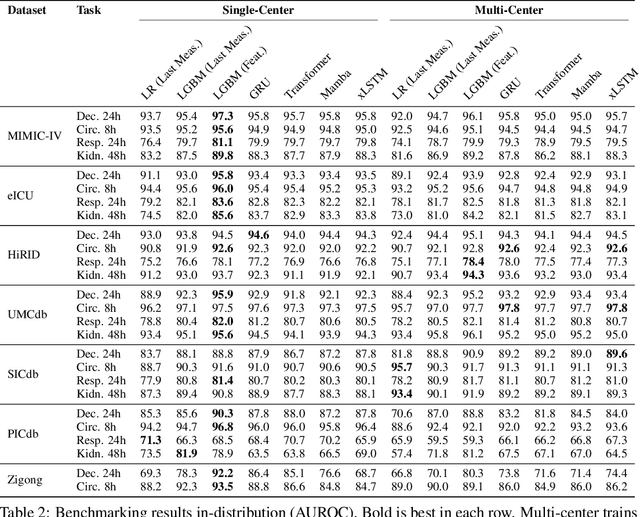
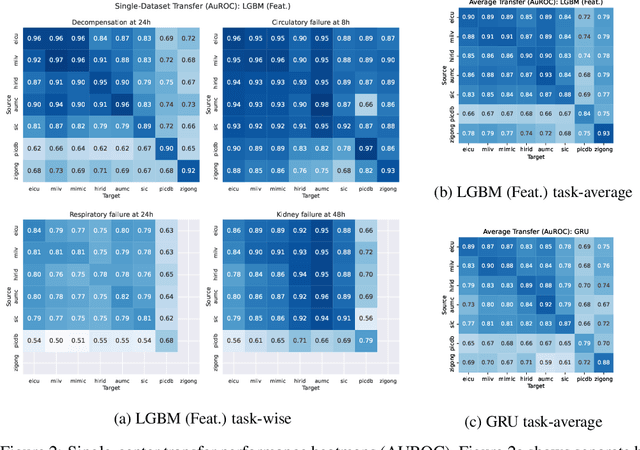
Abstract:Notable progress has been made in generalist medical large language models across various healthcare areas. However, large-scale modeling of in-hospital time series data - such as vital signs, lab results, and treatments in critical care - remains underexplored. Existing datasets are relatively small, but combining them can enhance patient diversity and improve model robustness. To effectively utilize these combined datasets for large-scale modeling, it is essential to address the distribution shifts caused by varying treatment policies, necessitating the harmonization of treatment variables across the different datasets. This work aims to establish a foundation for training large-scale multi-variate time series models on critical care data and to provide a benchmark for machine learning models in transfer learning across hospitals to study and address distribution shift challenges. We introduce a harmonized dataset for sequence modeling and transfer learning research, representing the first large-scale collection to include core treatment variables. Future plans involve expanding this dataset to support further advancements in transfer learning and the development of scalable, generalizable models for critical healthcare applications.
Generalizable Single-Source Cross-modality Medical Image Segmentation via Invariant Causal Mechanisms
Nov 07, 2024Abstract:Single-source domain generalization (SDG) aims to learn a model from a single source domain that can generalize well on unseen target domains. This is an important task in computer vision, particularly relevant to medical imaging where domain shifts are common. In this work, we consider a challenging yet practical setting: SDG for cross-modality medical image segmentation. We combine causality-inspired theoretical insights on learning domain-invariant representations with recent advancements in diffusion-based augmentation to improve generalization across diverse imaging modalities. Guided by the ``intervention-augmentation equivariant'' principle, we use controlled diffusion models (DMs) to simulate diverse imaging styles while preserving the content, leveraging rich generative priors in large-scale pretrained DMs to comprehensively perturb the multidimensional style variable. Extensive experiments on challenging cross-modality segmentation tasks demonstrate that our approach consistently outperforms state-of-the-art SDG methods across three distinct anatomies and imaging modalities. The source code is available at \href{https://github.com/ratschlab/ICMSeg}{https://github.com/ratschlab/ICMSeg}.
Uncertainty-Penalized Direct Preference Optimization
Oct 26, 2024



Abstract:Aligning Large Language Models (LLMs) to human preferences in content, style, and presentation is challenging, in part because preferences are varied, context-dependent, and sometimes inherently ambiguous. While successful, Reinforcement Learning from Human Feedback (RLHF) and Direct Preference Optimization (DPO) are prone to the issue of proxy reward overoptimization. Analysis of the DPO loss reveals a critical need for regularization for mislabeled or ambiguous preference pairs to avoid reward hacking. In this work, we develop a pessimistic framework for DPO by introducing preference uncertainty penalization schemes, inspired by offline reinforcement learning. The penalization serves as a correction to the loss which attenuates the loss gradient for uncertain samples. Evaluation of the methods is performed with GPT2 Medium on the Anthropic-HH dataset using a model ensemble to obtain uncertainty estimates, and shows improved overall performance compared to vanilla DPO, as well as better completions on prompts from high-uncertainty chosen/rejected responses.
Towards Dynamic Feature Acquisition on Medical Time Series by Maximizing Conditional Mutual Information
Jul 18, 2024



Abstract:Knowing which features of a multivariate time series to measure and when is a key task in medicine, wearables, and robotics. Better acquisition policies can reduce costs while maintaining or even improving the performance of downstream predictors. Inspired by the maximization of conditional mutual information, we propose an approach to train acquirers end-to-end using only the downstream loss. We show that our method outperforms random acquisition policy, matches a model with an unrestrained budget, but does not yet overtake a static acquisition strategy. We highlight the assumptions and outline avenues for future work.
Preference Elicitation for Offline Reinforcement Learning
Jun 26, 2024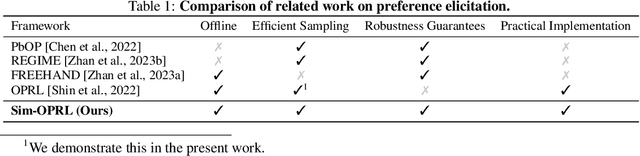
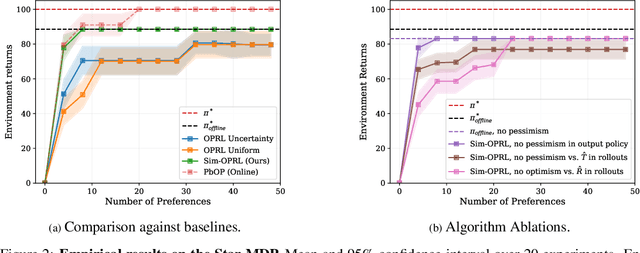

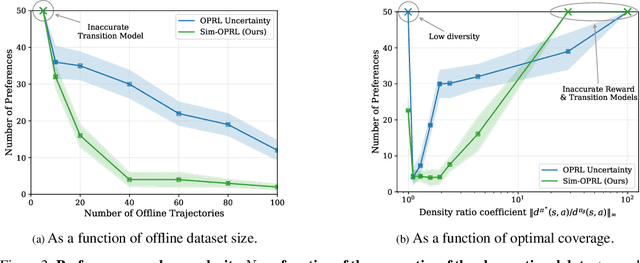
Abstract:Applying reinforcement learning (RL) to real-world problems is often made challenging by the inability to interact with the environment and the difficulty of designing reward functions. Offline RL addresses the first challenge by considering access to an offline dataset of environment interactions labeled by the reward function. In contrast, Preference-based RL does not assume access to the reward function and learns it from preferences, but typically requires an online interaction with the environment. We bridge the gap between these frameworks by exploring efficient methods for acquiring preference feedback in a fully offline setup. We propose Sim-OPRL, an offline preference-based reinforcement learning algorithm, which leverages a learned environment model to elicit preference feedback on simulated rollouts. Drawing on insights from both the offline RL and the preference-based RL literature, our algorithm employs a pessimistic approach for out-of-distribution data, and an optimistic approach for acquiring informative preferences about the optimal policy. We provide theoretical guarantees regarding the sample complexity of our approach, dependent on how well the offline data covers the optimal policy. Finally, we demonstrate the empirical performance of Sim-OPRL in different environments.
Multi-Modal Contrastive Learning for Online Clinical Time-Series Applications
Mar 27, 2024


Abstract:Electronic Health Record (EHR) datasets from Intensive Care Units (ICU) contain a diverse set of data modalities. While prior works have successfully leveraged multiple modalities in supervised settings, we apply advanced self-supervised multi-modal contrastive learning techniques to ICU data, specifically focusing on clinical notes and time-series for clinically relevant online prediction tasks. We introduce a loss function Multi-Modal Neighborhood Contrastive Loss (MM-NCL), a soft neighborhood function, and showcase the excellent linear probe and zero-shot performance of our approach.
Dynamic Survival Analysis for Early Event Prediction
Mar 19, 2024Abstract:This study advances Early Event Prediction (EEP) in healthcare through Dynamic Survival Analysis (DSA), offering a novel approach by integrating risk localization into alarm policies to enhance clinical event metrics. By adapting and evaluating DSA models against traditional EEP benchmarks, our research demonstrates their ability to match EEP models on a time-step level and significantly improve event-level metrics through a new alarm prioritization scheme (up to 11% AuPRC difference). This approach represents a significant step forward in predictive healthcare, providing a more nuanced and actionable framework for early event prediction and management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge