Khushbu Agarwal
Thinking Fast and Laterally: Multi-Agentic Approach for Reasoning about Uncertain Emerging Events
Dec 10, 2024Abstract:This paper introduces lateral thinking to implement System-2 reasoning capabilities in AI systems, focusing on anticipatory and causal reasoning under uncertainty. We present a framework for systematic generation and modeling of lateral thinking queries and evaluation datasets. We introduce Streaming Agentic Lateral Thinking (SALT), a multi-agent framework designed to process complex, low-specificity queries in streaming data environments. SALT implements lateral thinking-inspired System-2 reasoning through a dynamic communication structure between specialized agents. Our key insight is that lateral information flow across long-distance agent interactions, combined with fine-grained belief management, yields richer information contexts and enhanced reasoning. Preliminary quantitative and qualitative evaluations indicate SALT's potential to outperform single-agent systems in handling complex lateral reasoning tasks in a streaming environment.
ChemReasoner: Heuristic Search over a Large Language Model's Knowledge Space using Quantum-Chemical Feedback
Feb 21, 2024



Abstract:The discovery of new catalysts is essential for the design of new and more efficient chemical processes in order to transition to a sustainable future. We introduce an AI-guided computational screening framework unifying linguistic reasoning with quantum-chemistry based feedback from 3D atomistic representations. Our approach formulates catalyst discovery as an uncertain environment where an agent actively searches for highly effective catalysts via the iterative combination of large language model (LLM)-derived hypotheses and atomistic graph neural network (GNN)-derived feedback. Identified catalysts in intermediate search steps undergo structural evaluation based on spatial orientation, reaction pathways, and stability. Scoring functions based on adsorption energies and barriers steer the exploration in the LLM's knowledge space toward energetically favorable, high-efficiency catalysts. We introduce planning methods that automatically guide the exploration without human input, providing competitive performance against expert-enumerated chemical descriptor-based implementations. By integrating language-guided reasoning with computational chemistry feedback, our work pioneers AI-accelerated, trustworthy catalyst discovery.
GLaM: Fine-Tuning Large Language Models for Domain Knowledge Graph Alignment via Neighborhood Partitioning and Generative Subgraph Encoding
Feb 16, 2024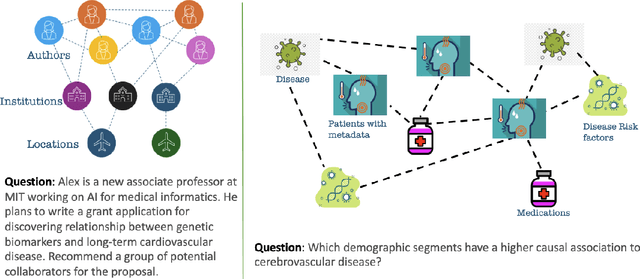
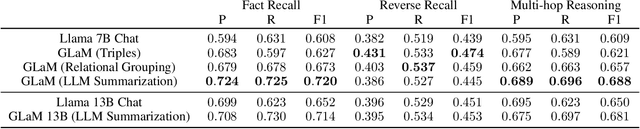
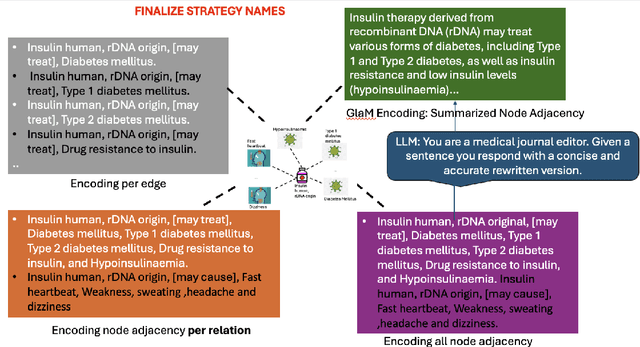
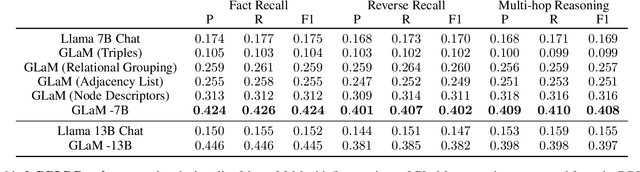
Abstract:Integrating large language models (LLMs) with knowledge graphs derived from domain-specific data represents an important advancement towards more powerful and factual reasoning. As these models grow more capable, it is crucial to enable them to perform multi-step inferences over real-world knowledge graphs while minimizing hallucination. While large language models excel at conversation and text generation, their ability to reason over domain-specialized graphs of interconnected entities remains limited. For example, can we query a LLM to identify the optimal contact in a professional network for a specific goal, based on relationships and attributes in a private database? The answer is no--such capabilities lie beyond current methods. However, this question underscores a critical technical gap that must be addressed. Many high-value applications in areas such as science, security, and e-commerce rely on proprietary knowledge graphs encoding unique structures, relationships, and logical constraints. We introduce a fine-tuning framework for developing Graph-aligned LAnguage Models (GLaM) that transforms a knowledge graph into an alternate text representation with labeled question-answer pairs. We demonstrate that grounding the models in specific graph-based knowledge expands the models' capacity for structure-based reasoning. Our methodology leverages the large-language model's generative capabilities to create the dataset and proposes an efficient alternate to retrieval-augmented generation styled methods.
Pseudo-Poincaré: A Unification Framework for Euclidean and Hyperbolic Graph Neural Networks
Jun 09, 2022



Abstract:Hyperbolic neural networks have recently gained significant attention due to their promising results on several graph problems including node classification and link prediction. The primary reason for this success is the effectiveness of the hyperbolic space in capturing the inherent hierarchy of graph datasets. However, they are limited in terms of generalization, scalability, and have inferior performance when it comes to non-hierarchical datasets. In this paper, we take a completely orthogonal perspective for modeling hyperbolic networks. We use Poincar\'e disk to model the hyperbolic geometry and also treat it as if the disk itself is a tangent space at origin. This enables us to replace non-scalable M\"obius gyrovector operations with an Euclidean approximation, and thus simplifying the entire hyperbolic model to a Euclidean model cascaded with a hyperbolic normalization function. Our approach does not adhere to M\"obius math, yet it still works in the Riemannian manifold, hence we call it Pseudo-Poincar\'e framework. We applied our non-linear hyperbolic normalization to the current state-of-the-art homogeneous and multi-relational graph networks and demonstrate significant improvements in performance compared to both Euclidean and hyperbolic counterparts. The primary impact of this work lies in its ability to capture hierarchical features in the Euclidean space, and thus, can replace hyperbolic networks without loss in performance metrics while simultaneously leveraging the power of Euclidean networks such as interpretability and efficient execution of various model components.
Attention-based Aspect Reasoning for Knowledge Base Question Answering on Clinical Notes
Aug 01, 2021

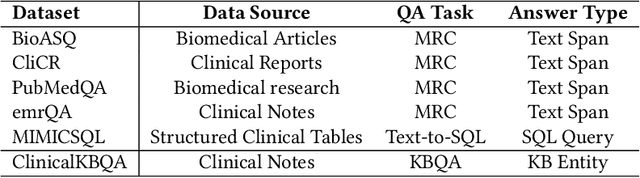
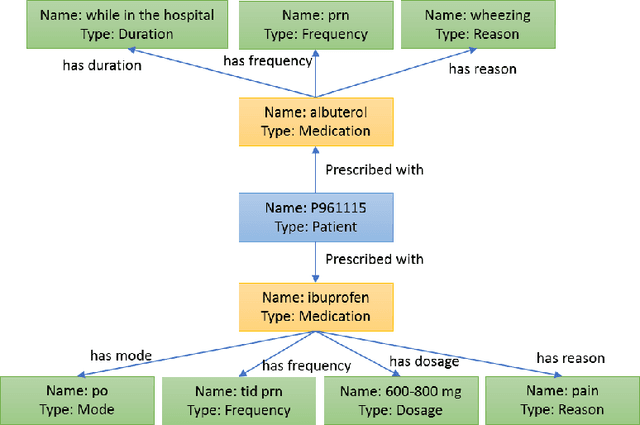
Abstract:Question Answering (QA) in clinical notes has gained a lot of attention in the past few years. Existing machine reading comprehension approaches in clinical domain can only handle questions about a single block of clinical texts and fail to retrieve information about different patients and clinical notes. To handle more complex questions, we aim at creating knowledge base from clinical notes to link different patients and clinical notes, and performing knowledge base question answering (KBQA). Based on the expert annotations in n2c2, we first created the ClinicalKBQA dataset that includes 8,952 QA pairs and covers questions about seven medical topics through 322 question templates. Then, we proposed an attention-based aspect reasoning (AAR) method for KBQA and investigated the impact of different aspects of answers (e.g., entity, type, path, and context) for prediction. The AAR method achieves better performance due to the well-designed encoder and attention mechanism. In the experiments, we find that both aspects, type and path, enable the model to identify answers satisfying the general conditions and produce lower precision and higher recall. On the other hand, the aspects, entity and context, limit the answers by node-specific information and lead to higher precision and lower recall.
Self-Supervised Learning of Contextual Embeddings for Link Prediction in Heterogeneous Networks
Aug 23, 2020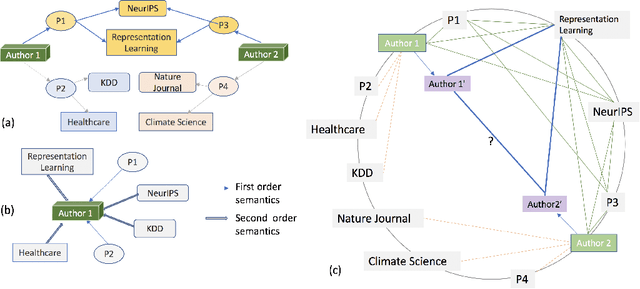

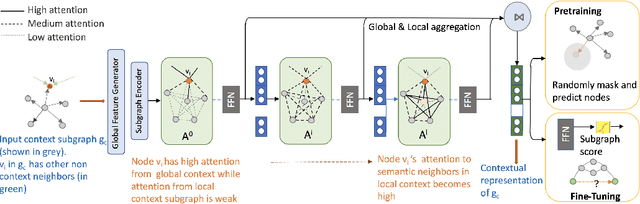

Abstract:Representation learning methods for heterogeneous networks produce a low-dimensional vector embedding for each node that is typically fixed for all tasks involving the node. Many of the existing methods focus on obtaining a static vector representation for a node in a way that is agnostic to the downstream application where it is being used. In practice, however, downstream tasks such as link prediction require specific contextual information that can be extracted from the subgraphs related to the nodes provided as input to the task. To tackle this challenge, we develop SLiCE, a framework bridging static representation learning methods using global information from the entire graph with localized attention driven mechanisms to learn contextual node representations. We first pre-train our model in a self-supervised manner by introducing higher-order semantic associations and masking nodes, and then fine-tune our model for a specific link prediction task. Instead of training node representations by aggregating information from all semantic neighbors connected via metapaths, we automatically learn the composition of different metapaths that characterize the context for a specific task without the need for any pre-defined metapaths. SLiCE significantly outperforms both static and contextual embedding learning methods on several publicly available benchmark network datasets. We also interpret the semantic association matrix and provide its utility and relevance in making successful link predictions between heterogeneous nodes in the network.
Snomed2Vec: Random Walk and Poincaré Embeddings of a Clinical Knowledge Base for Healthcare Analytics
Jul 19, 2019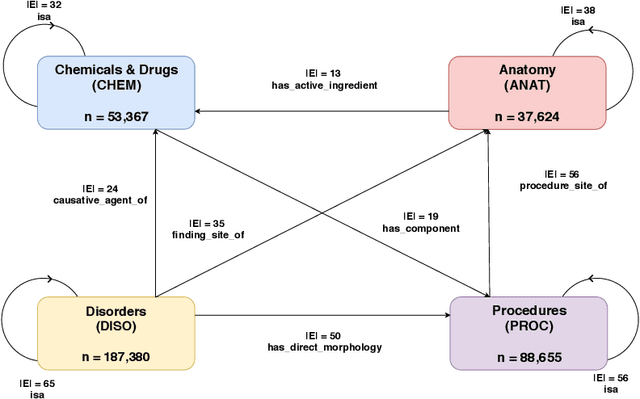
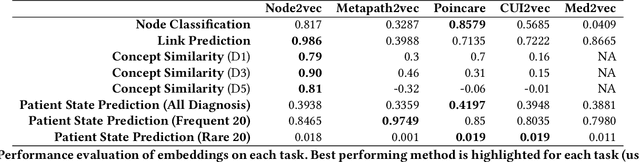
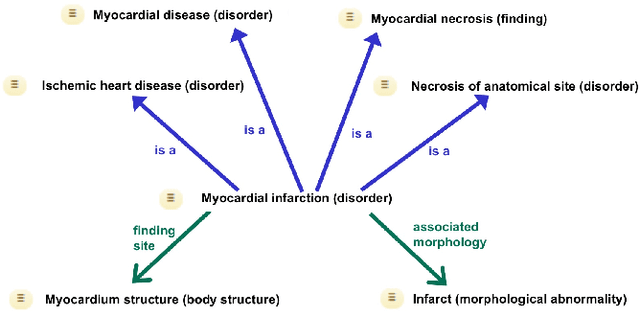
Abstract:Representation learning methods that transform encoded data (e.g., diagnosis and drug codes) into continuous vector spaces (i.e., vector embeddings) are critical for the application of deep learning in healthcare. Initial work in this area explored the use of variants of the word2vec algorithm to learn embeddings for medical concepts from electronic health records or medical claims datasets. We propose learning embeddings for medical concepts by using graph-based representation learning methods on SNOMED-CT, a widely popular knowledge graph in the healthcare domain with numerous operational and research applications. Current work presents an empirical analysis of various embedding methods, including the evaluation of their performance on multiple tasks of biomedical relevance (node classification, link prediction, and patient state prediction). Our results show that concept embeddings derived from the SNOMED-CT knowledge graph significantly outperform state-of-the-art embeddings, showing 5-6x improvement in ``concept similarity" and 6-20\% improvement in patient diagnosis.
Trust from the past: Bayesian Personalized Ranking based Link Prediction in Knowledge Graphs
Feb 15, 2016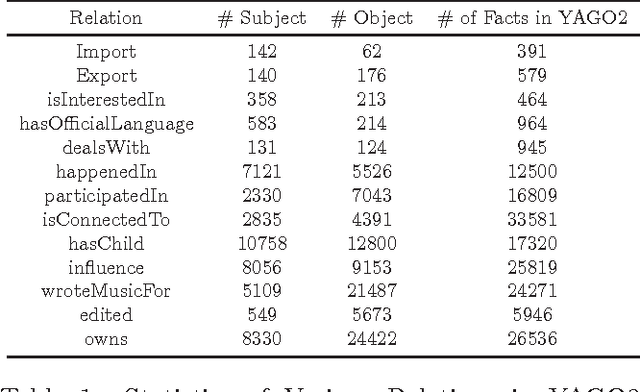
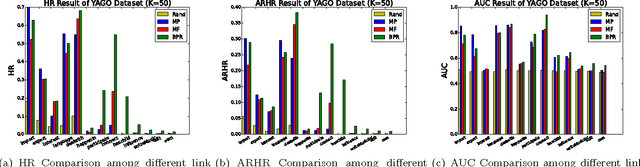

Abstract:Link prediction, or predicting the likelihood of a link in a knowledge graph based on its existing state is a key research task. It differs from a traditional link prediction task in that the links in a knowledge graph are categorized into different predicates and the link prediction performance of different predicates in a knowledge graph generally varies widely. In this work, we propose a latent feature embedding based link prediction model which considers the prediction task for each predicate disjointly. To learn the model parameters it utilizes a Bayesian personalized ranking based optimization technique. Experimental results on large-scale knowledge bases such as YAGO2 show that our link prediction approach achieves substantially higher performance than several state-of-art approaches. We also show that for a given predicate the topological properties of the knowledge graph induced by the given predicate edges are key indicators of the link prediction performance of that predicate in the knowledge graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge