Attention-based Aspect Reasoning for Knowledge Base Question Answering on Clinical Notes
Paper and Code
Aug 01, 2021

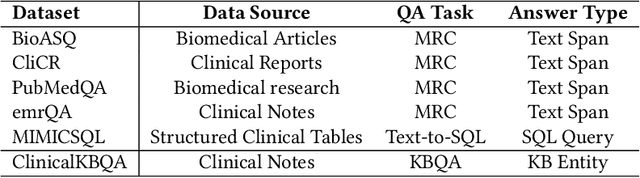
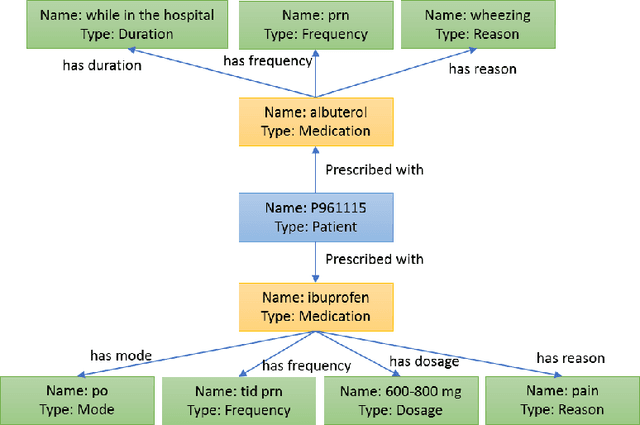
Question Answering (QA) in clinical notes has gained a lot of attention in the past few years. Existing machine reading comprehension approaches in clinical domain can only handle questions about a single block of clinical texts and fail to retrieve information about different patients and clinical notes. To handle more complex questions, we aim at creating knowledge base from clinical notes to link different patients and clinical notes, and performing knowledge base question answering (KBQA). Based on the expert annotations in n2c2, we first created the ClinicalKBQA dataset that includes 8,952 QA pairs and covers questions about seven medical topics through 322 question templates. Then, we proposed an attention-based aspect reasoning (AAR) method for KBQA and investigated the impact of different aspects of answers (e.g., entity, type, path, and context) for prediction. The AAR method achieves better performance due to the well-designed encoder and attention mechanism. In the experiments, we find that both aspects, type and path, enable the model to identify answers satisfying the general conditions and produce lower precision and higher recall. On the other hand, the aspects, entity and context, limit the answers by node-specific information and lead to higher precision and lower recall.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge