Jorge Bernal
Enhancing Image Classification in Small and Unbalanced Datasets through Synthetic Data Augmentation
Sep 16, 2024Abstract:Accurate and robust medical image classification is a challenging task, especially in application domains where available annotated datasets are small and present high imbalance between target classes. Considering that data acquisition is not always feasible, especially for underrepresented classes, our approach introduces a novel synthetic augmentation strategy using class-specific Variational Autoencoders (VAEs) and latent space interpolation to improve discrimination capabilities. By generating realistic, varied synthetic data that fills feature space gaps, we address issues of data scarcity and class imbalance. The method presented in this paper relies on the interpolation of latent representations within each class, thus enriching the training set and improving the model's generalizability and diagnostic accuracy. The proposed strategy was tested in a small dataset of 321 images created to train and validate an automatic method for assessing the quality of cleanliness of esophagogastroduodenoscopy images. By combining real and synthetic data, an increase of over 18\% in the accuracy of the most challenging underrepresented class was observed. The proposed strategy not only benefited the underrepresented class but also led to a general improvement in other metrics, including a 6\% increase in global accuracy and precision.
Why is the winner the best?
Mar 30, 2023
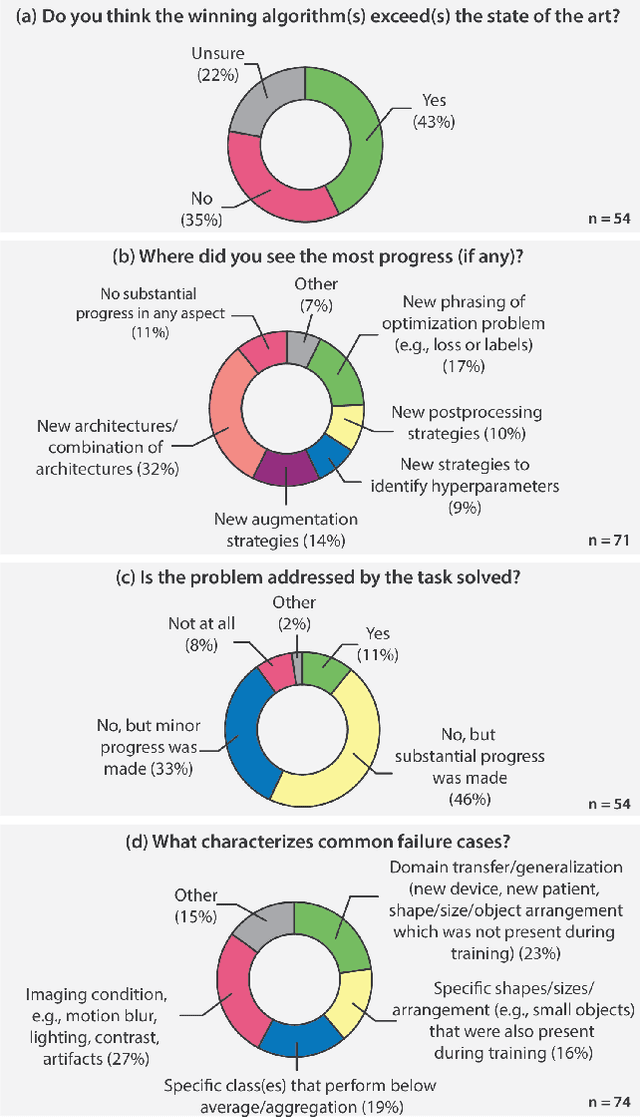
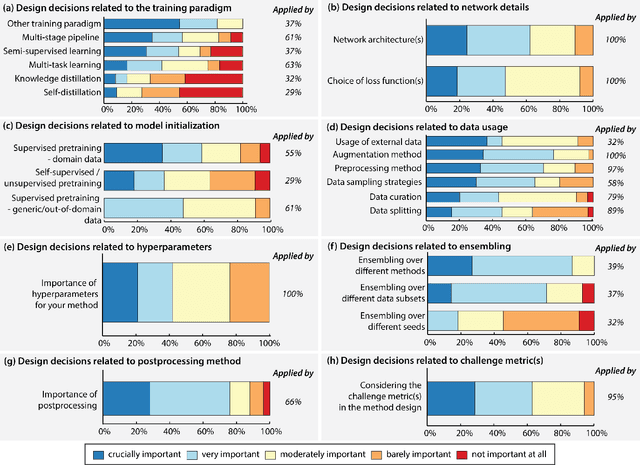
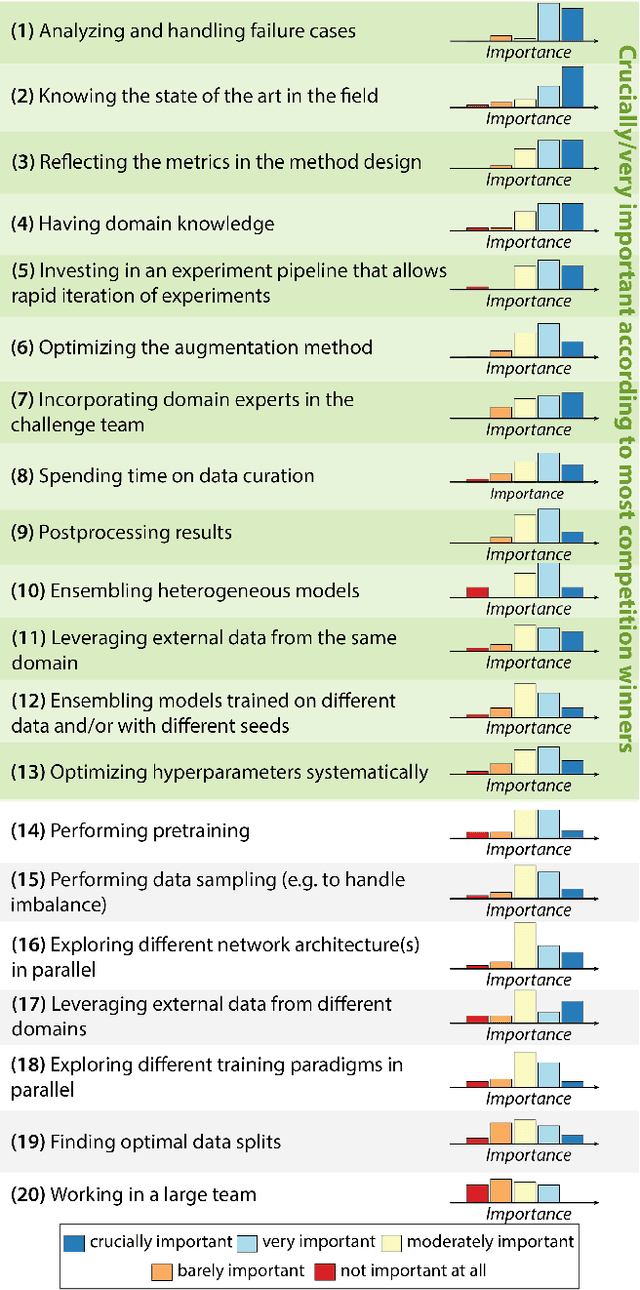
Abstract:International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
Biomedical image analysis competitions: The state of current participation practice
Dec 16, 2022Abstract:The number of international benchmarking competitions is steadily increasing in various fields of machine learning (ML) research and practice. So far, however, little is known about the common practice as well as bottlenecks faced by the community in tackling the research questions posed. To shed light on the status quo of algorithm development in the specific field of biomedical imaging analysis, we designed an international survey that was issued to all participants of challenges conducted in conjunction with the IEEE ISBI 2021 and MICCAI 2021 conferences (80 competitions in total). The survey covered participants' expertise and working environments, their chosen strategies, as well as algorithm characteristics. A median of 72% challenge participants took part in the survey. According to our results, knowledge exchange was the primary incentive (70%) for participation, while the reception of prize money played only a minor role (16%). While a median of 80 working hours was spent on method development, a large portion of participants stated that they did not have enough time for method development (32%). 25% perceived the infrastructure to be a bottleneck. Overall, 94% of all solutions were deep learning-based. Of these, 84% were based on standard architectures. 43% of the respondents reported that the data samples (e.g., images) were too large to be processed at once. This was most commonly addressed by patch-based training (69%), downsampling (37%), and solving 3D analysis tasks as a series of 2D tasks. K-fold cross-validation on the training set was performed by only 37% of the participants and only 50% of the participants performed ensembling based on multiple identical models (61%) or heterogeneous models (39%). 48% of the respondents applied postprocessing steps.
Towards a Computed-Aided Diagnosis System in Colonoscopy: Automatic Polyp Segmentation Using Convolution Neural Networks
Jan 15, 2021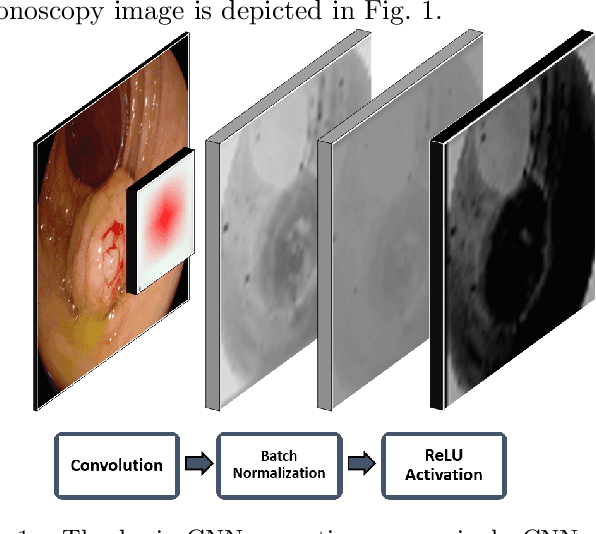
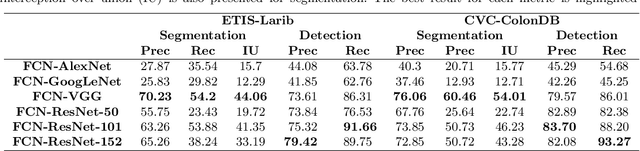
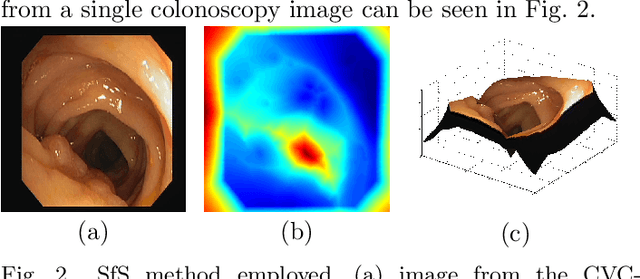
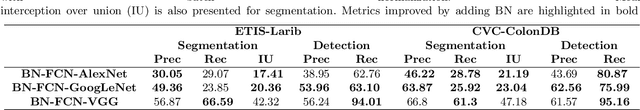
Abstract:Early diagnosis is essential for the successful treatment of bowel cancers including colorectal cancer (CRC) and capsule endoscopic imaging with robotic actuation can be a valuable diagnostic tool when combined with automated image analysis. We present a deep learning rooted detection and segmentation framework for recognizing lesions in colonoscopy and capsule endoscopy images. We restructure established convolution architectures, such as VGG and ResNets, by converting them into fully-connected convolution networks (FCNs), fine-tune them and study their capabilities for polyp segmentation and detection. We additionally use Shape from-Shading (SfS) to recover depth and provide a richer representation of the tissue's structure in colonoscopy images. Depth is incorporated into our network models as an additional input channel to the RGB information and we demonstrate that the resulting network yields improved performance. Our networks are tested on publicly available datasets and the most accurate segmentation model achieved a mean segmentation IU of 47.78% and 56.95% on the ETIS-Larib and CVC-Colon datasets, respectively. For polyp detection, the top performing models we propose surpass the current state of the art with detection recalls superior to 90% for all datasets tested. To our knowledge, we present the first work to use FCNs for polyp segmentation in addition to proposing a novel combination of SfS and RGB that boosts performance
* 10 pages, 6 figures
A Benchmark for Endoluminal Scene Segmentation of Colonoscopy Images
Dec 02, 2016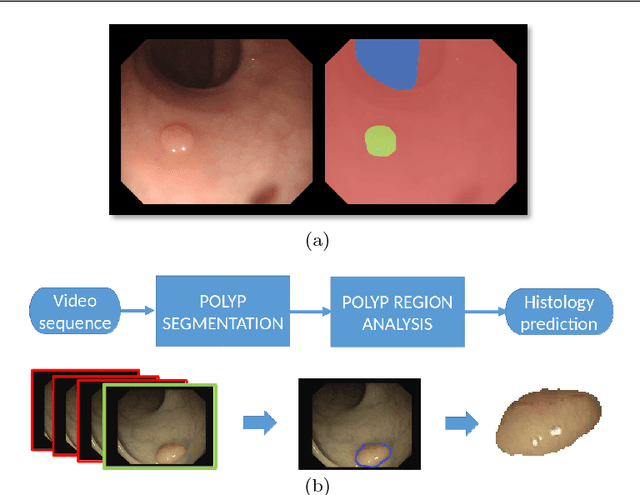
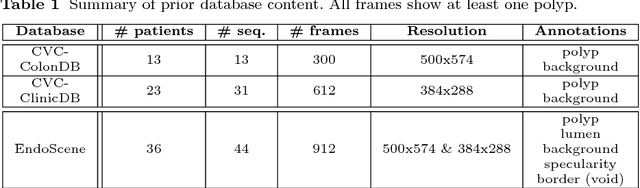

Abstract:Colorectal cancer (CRC) is the third cause of cancer death worldwide. Currently, the standard approach to reduce CRC-related mortality is to perform regular screening in search for polyps and colonoscopy is the screening tool of choice. The main limitations of this screening procedure are polyp miss-rate and inability to perform visual assessment of polyp malignancy. These drawbacks can be reduced by designing Decision Support Systems (DSS) aiming to help clinicians in the different stages of the procedure by providing endoluminal scene segmentation. Thus, in this paper, we introduce an extended benchmark of colonoscopy image, with the hope of establishing a new strong benchmark for colonoscopy image analysis research. We provide new baselines on this dataset by training standard fully convolutional networks (FCN) for semantic segmentation and significantly outperforming, without any further post-processing, prior results in endoluminal scene segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge