Jithendaraa Subramanian
Context is Key: A Benchmark for Forecasting with Essential Textual Information
Oct 24, 2024



Abstract:Forecasting is a critical task in decision making across various domains. While numerical data provides a foundation, it often lacks crucial context necessary for accurate predictions. Human forecasters frequently rely on additional information, such as background knowledge or constraints, which can be efficiently communicated through natural language. However, the ability of existing forecasting models to effectively integrate this textual information remains an open question. To address this, we introduce "Context is Key" (CiK), a time series forecasting benchmark that pairs numerical data with diverse types of carefully crafted textual context, requiring models to integrate both modalities. We evaluate a range of approaches, including statistical models, time series foundation models, and LLM-based forecasters, and propose a simple yet effective LLM prompting method that outperforms all other tested methods on our benchmark. Our experiments highlight the importance of incorporating contextual information, demonstrate surprising performance when using LLM-based forecasting models, and also reveal some of their critical shortcomings. By presenting this benchmark, we aim to advance multimodal forecasting, promoting models that are both accurate and accessible to decision-makers with varied technical expertise. The benchmark can be visualized at https://servicenow.github.io/context-is-key-forecasting/v0/ .
Reinforcement Learning for Sequence Design Leveraging Protein Language Models
Jul 03, 2024



Abstract:Protein sequence design, determined by amino acid sequences, are essential to protein engineering problems in drug discovery. Prior approaches have resorted to evolutionary strategies or Monte-Carlo methods for protein design, but often fail to exploit the structure of the combinatorial search space, to generalize to unseen sequences. In the context of discrete black box optimization over large search spaces, learning a mutation policy to generate novel sequences with reinforcement learning is appealing. Recent advances in protein language models (PLMs) trained on large corpora of protein sequences offer a potential solution to this problem by scoring proteins according to their biological plausibility (such as the TM-score). In this work, we propose to use PLMs as a reward function to generate new sequences. Yet the PLM can be computationally expensive to query due to its large size. To this end, we propose an alternative paradigm where optimization can be performed on scores from a smaller proxy model that is periodically finetuned, jointly while learning the mutation policy. We perform extensive experiments on various sequence lengths to benchmark RL-based approaches, and provide comprehensive evaluations along biological plausibility and diversity of the protein. Our experimental results include favorable evaluations of the proposed sequences, along with high diversity scores, demonstrating that RL is a strong candidate for biological sequence design. Finally, we provide a modular open source implementation can be easily integrated in most RL training loops, with support for replacing the reward model with other PLMs, to spur further research in this domain. The code for all experiments is provided in the supplementary material.
Joint Bayesian Inference of Graphical Structure and Parameters with a Single Generative Flow Network
May 30, 2023



Abstract:Generative Flow Networks (GFlowNets), a class of generative models over discrete and structured sample spaces, have been previously applied to the problem of inferring the marginal posterior distribution over the directed acyclic graph (DAG) of a Bayesian Network, given a dataset of observations. Based on recent advances extending this framework to non-discrete sample spaces, we propose in this paper to approximate the joint posterior over not only the structure of a Bayesian Network, but also the parameters of its conditional probability distributions. We use a single GFlowNet whose sampling policy follows a two-phase process: the DAG is first generated sequentially one edge at a time, and then the corresponding parameters are picked once the full structure is known. Since the parameters are included in the posterior distribution, this leaves more flexibility for the local probability models of the Bayesian Network, making our approach applicable even to non-linear models parametrized by neural networks. We show that our method, called JSP-GFN, offers an accurate approximation of the joint posterior, while comparing favorably against existing methods on both simulated and real data.
Bayesian learning of Causal Structure and Mechanisms with GFlowNets and Variational Bayes
Nov 04, 2022



Abstract:Bayesian causal structure learning aims to learn a posterior distribution over directed acyclic graphs (DAGs), and the mechanisms that define the relationship between parent and child variables. By taking a Bayesian approach, it is possible to reason about the uncertainty of the causal model. The notion of modelling the uncertainty over models is particularly crucial for causal structure learning since the model could be unidentifiable when given only a finite amount of observational data. In this paper, we introduce a novel method to jointly learn the structure and mechanisms of the causal model using Variational Bayes, which we call Variational Bayes-DAG-GFlowNet (VBG). We extend the method of Bayesian causal structure learning using GFlowNets to learn not only the posterior distribution over the structure, but also the parameters of a linear-Gaussian model. Our results on simulated data suggest that VBG is competitive against several baselines in modelling the posterior over DAGs and mechanisms, while offering several advantages over existing methods, including the guarantee to sample acyclic graphs, and the flexibility to generalize to non-linear causal mechanisms.
Learning Latent Structural Causal Models
Oct 24, 2022



Abstract:Causal learning has long concerned itself with the accurate recovery of underlying causal mechanisms. Such causal modelling enables better explanations of out-of-distribution data. Prior works on causal learning assume that the high-level causal variables are given. However, in machine learning tasks, one often operates on low-level data like image pixels or high-dimensional vectors. In such settings, the entire Structural Causal Model (SCM) -- structure, parameters, \textit{and} high-level causal variables -- is unobserved and needs to be learnt from low-level data. We treat this problem as Bayesian inference of the latent SCM, given low-level data. For linear Gaussian additive noise SCMs, we present a tractable approximate inference method which performs joint inference over the causal variables, structure and parameters of the latent SCM from random, known interventions. Experiments are performed on synthetic datasets and a causally generated image dataset to demonstrate the efficacy of our approach. We also perform image generation from unseen interventions, thereby verifying out of distribution generalization for the proposed causal model.
Latent Variable Models for Bayesian Causal Discovery
Jul 12, 2022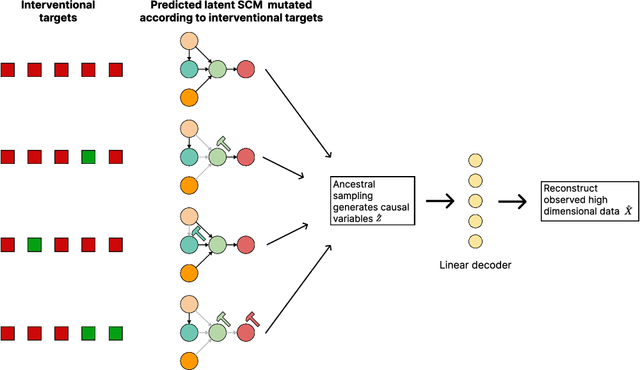
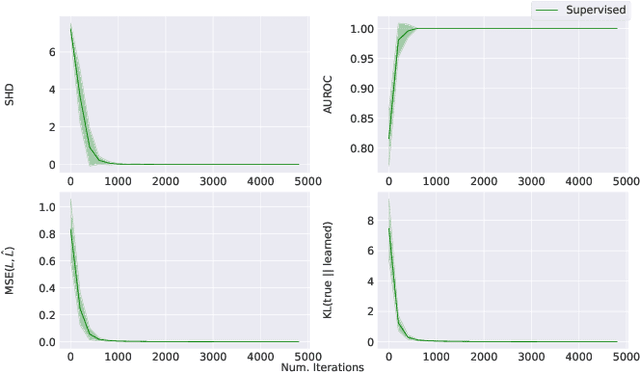
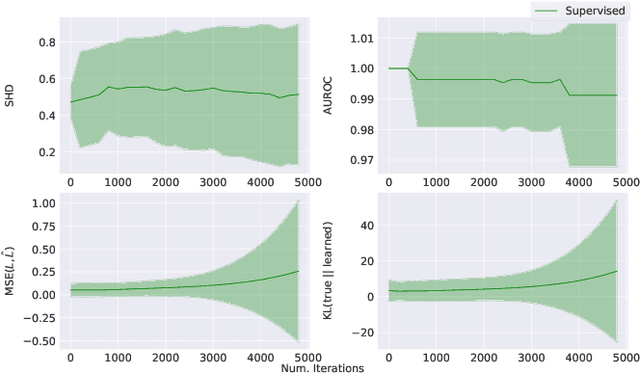
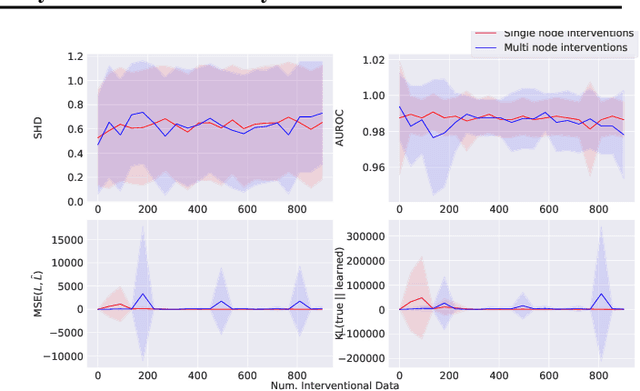
Abstract:Learning predictors that do not rely on spurious correlations involves building causal representations. However, learning such a representation is very challenging. We, therefore, formulate the problem of learning a causal representation from high dimensional data and study causal recovery with synthetic data. This work introduces a latent variable decoder model, Decoder BCD, for Bayesian causal discovery and performs experiments in mildly supervised and unsupervised settings. We present a series of synthetic experiments to characterize important factors for causal discovery and show that using known intervention targets as labels helps in unsupervised Bayesian inference over structure and parameters of linear Gaussian additive noise latent structural causal models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge