Jiankai Tang
Non-Contact Health Monitoring During Daily Personal Care Routines
Jun 11, 2025Abstract:Remote photoplethysmography (rPPG) enables non-contact, continuous monitoring of physiological signals and offers a practical alternative to traditional health sensing methods. Although rPPG is promising for daily health monitoring, its application in long-term personal care scenarios, such as mirror-facing routines in high-altitude environments, remains challenging due to ambient lighting variations, frequent occlusions from hand movements, and dynamic facial postures. To address these challenges, we present LADH (Long-term Altitude Daily Health), the first long-term rPPG dataset containing 240 synchronized RGB and infrared (IR) facial videos from 21 participants across five common personal care scenarios, along with ground-truth PPG, respiration, and blood oxygen signals. Our experiments demonstrate that combining RGB and IR video inputs improves the accuracy and robustness of non-contact physiological monitoring, achieving a mean absolute error (MAE) of 4.99 BPM in heart rate estimation. Furthermore, we find that multi-task learning enhances performance across multiple physiological indicators simultaneously. Dataset and code are open at https://github.com/McJackTang/FusionVitals.
A Dataset and Toolkit for Multiparameter Cardiovascular Physiology Sensing on Rings
May 08, 2025



Abstract:Smart rings offer a convenient way to continuously and unobtrusively monitor cardiovascular physiological signals. However, a gap remains between the ring hardware and reliable methods for estimating cardiovascular parameters, partly due to the lack of publicly available datasets and standardized analysis tools. In this work, we present $\tau$-Ring, the first open-source ring-based dataset designed for cardiovascular physiological sensing. The dataset comprises photoplethysmography signals (infrared and red channels) and 3-axis accelerometer data collected from two rings (reflective and transmissive optical paths), with 28.21 hours of raw data from 34 subjects across seven activities. $\tau$-Ring encompasses both stationary and motion scenarios, as well as stimulus-evoked abnormal physiological states, annotated with four ground-truth labels: heart rate, respiratory rate, oxygen saturation, and blood pressure. Using our proposed RingTool toolkit, we evaluated three widely-used physics-based methods and four cutting-edge deep learning approaches. Our results show superior performance compared to commercial rings, achieving best MAE values of 5.18 BPM for heart rate, 2.98 BPM for respiratory rate, 3.22\% for oxygen saturation, and 13.33/7.56 mmHg for systolic/diastolic blood pressure estimation. The open-sourced dataset and toolkit aim to foster further research and community-driven advances in ring-based cardiovascular health sensing.
Memory-efficient Low-latency Remote Photoplethysmography through Temporal-Spatial State Space Duality
Apr 02, 2025Abstract:Remote photoplethysmography (rPPG), enabling non-contact physiological monitoring through facial light reflection analysis, faces critical computational bottlenecks as deep learning introduces performance gains at the cost of prohibitive resource demands. This paper proposes ME-rPPG, a memory-efficient algorithm built on temporal-spatial state space duality, which resolves the trilemma of model scalability, cross-dataset generalization, and real-time constraints. Leveraging a transferable state space, ME-rPPG efficiently captures subtle periodic variations across facial frames while maintaining minimal computational overhead, enabling training on extended video sequences and supporting low-latency inference. Achieving cross-dataset MAEs of 5.38 (MMPD), 0.70 (VitalVideo), and 0.25 (PURE), ME-rPPG outperforms all baselines with improvements ranging from 21.3% to 60.2%. Our solution enables real-time inference with only 3.6 MB memory usage and 9.46 ms latency -- surpassing existing methods by 19.5%-49.7% accuracy and 43.2% user satisfaction gains in real-world deployments. The code and demos are released for reproducibility on https://github.com/Health-HCI-Group/ME-rPPG-demo.
Exploring Reliable PPG Authentication on Smartwatches in Daily Scenarios
Mar 31, 2025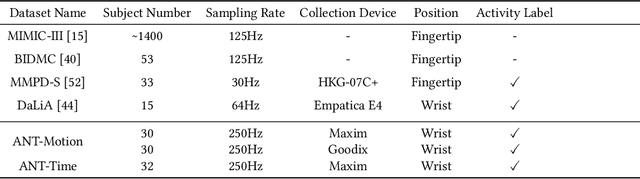
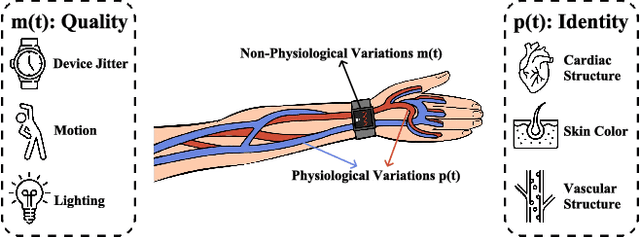
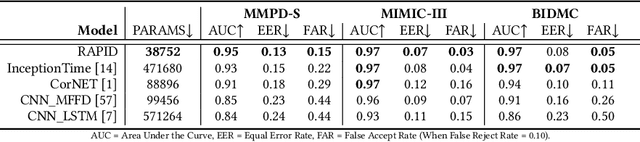
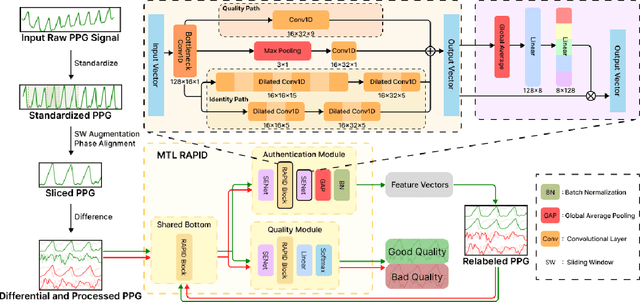
Abstract:Photoplethysmography (PPG) Sensors, widely deployed in smartwatches, offer a simple and non-invasive authentication approach for daily use. However, PPG authentication faces reliability issues due to motion artifacts from physical activity and physiological variability over time. To address these challenges, we propose MTL-RAPID, an efficient and reliable PPG authentication model, that employs a multitask joint training strategy, simultaneously assessing signal quality and verifying user identity. The joint optimization of these two tasks in MTL-RAPID results in a structure that outperforms models trained on individual tasks separately, achieving stronger performance with fewer parameters. In our comprehensive user studies regarding motion artifacts (N = 30), time variations (N = 32), and user preferences (N = 16), MTL-RAPID achieves a best AUC of 99.2\% and an EER of 3.5\%, outperforming existing baselines. We opensource our PPG authentication dataset along with the MTL-RAPID model to facilitate future research on GitHub.
A Plug-and-Play Temporal Normalization Module for Robust Remote Photoplethysmography
Nov 22, 2024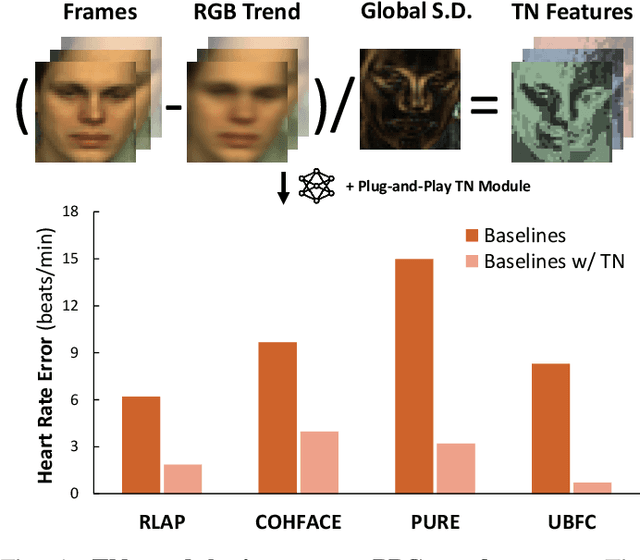
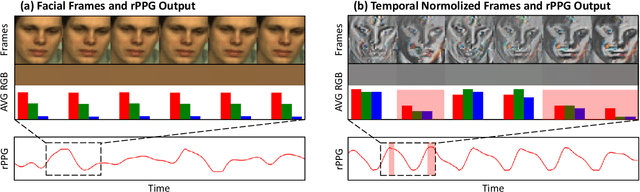
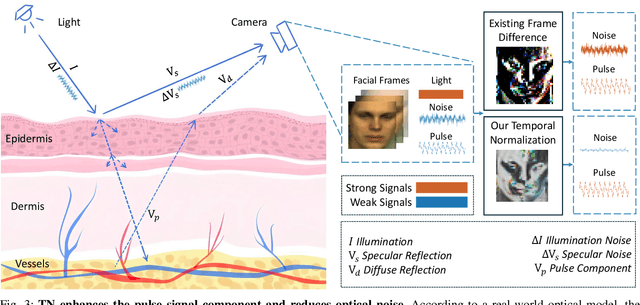
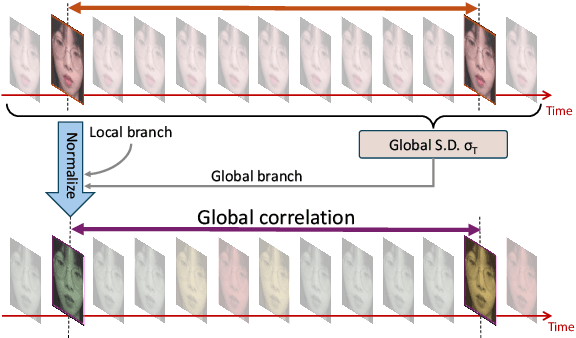
Abstract:Remote photoplethysmography (rPPG) extracts PPG signals from subtle color changes in facial videos, showing strong potential for health applications. However, most rPPG methods rely on intensity differences between consecutive frames, missing long-term signal variations affected by motion or lighting artifacts, which reduces accuracy. This paper introduces Temporal Normalization (TN), a flexible plug-and-play module compatible with any end-to-end rPPG network architecture. By capturing long-term temporally normalized features following detrending, TN effectively mitigates motion and lighting artifacts, significantly boosting the rPPG prediction performance. When integrated into four state-of-the-art rPPG methods, TN delivered performance improvements ranging from 34.3% to 94.2% in heart rate measurement tasks across four widely-used datasets. Notably, TN showed even greater performance gains in smaller models. We further discuss and provide insights into the mechanisms behind TN's effectiveness.
Summit Vitals: Multi-Camera and Multi-Signal Biosensing at High Altitudes
Sep 28, 2024Abstract:Video photoplethysmography (vPPG) is an emerging method for non-invasive and convenient measurement of physiological signals, utilizing two primary approaches: remote video PPG (rPPG) and contact video PPG (cPPG). Monitoring vitals in high-altitude environments, where heart rates tend to increase and blood oxygen levels often decrease, presents significant challenges. To address these issues, we introduce the SUMS dataset comprising 80 synchronized non-contact facial and contact finger videos from 10 subjects during exercise and oxygen recovery scenarios, capturing PPG, respiration rate (RR), and SpO2. This dataset is designed to validate video vitals estimation algorithms and compare facial rPPG with finger cPPG. Additionally, fusing videos from different positions (i.e., face and finger) reduces the mean absolute error (MAE) of SpO2 predictions by 7.6\% and 10.6\% compared to only face and only finger, respectively. In cross-subject evaluation, we achieve an MAE of less than 0.5 BPM for HR estimation and 2.5\% for SpO2 estimation, demonstrating the precision of our multi-camera fusion techniques. Our findings suggest that simultaneous training on multiple indicators, such as PPG and blood oxygen, can reduce MAE in SpO2 estimation by 17.8\%.
Camera-Based Remote Physiology Sensing for Hundreds of Subjects Across Skin Tones
Apr 07, 2024



Abstract:Remote photoplethysmography (rPPG) emerges as a promising method for non-invasive, convenient measurement of vital signs, utilizing the widespread presence of cameras. Despite advancements, existing datasets fall short in terms of size and diversity, limiting comprehensive evaluation under diverse conditions. This paper presents an in-depth analysis of the VitalVideo dataset, the largest real-world rPPG dataset to date, encompassing 893 subjects and 6 Fitzpatrick skin tones. Our experimentation with six unsupervised methods and three supervised models demonstrates that datasets comprising a few hundred subjects(i.e., 300 for UBFC-rPPG, 500 for PURE, and 700 for MMPD-Simple) are sufficient for effective rPPG model training. Our findings highlight the importance of diversity and consistency in skin tones for precise performance evaluation across different datasets.
Spiking-PhysFormer: Camera-Based Remote Photoplethysmography with Parallel Spike-driven Transformer
Feb 09, 2024Abstract:Artificial neural networks (ANNs) can help camera-based remote photoplethysmography (rPPG) in measuring cardiac activity and physiological signals from facial videos, such as pulse wave, heart rate and respiration rate with better accuracy. However, most existing ANN-based methods require substantial computing resources, which poses challenges for effective deployment on mobile devices. Spiking neural networks (SNNs), on the other hand, hold immense potential for energy-efficient deep learning owing to their binary and event-driven architecture. To the best of our knowledge, we are the first to introduce SNNs into the realm of rPPG, proposing a hybrid neural network (HNN) model, the Spiking-PhysFormer, aimed at reducing power consumption. Specifically, the proposed Spiking-PhyFormer consists of an ANN-based patch embedding block, SNN-based transformer blocks, and an ANN-based predictor head. First, to simplify the transformer block while preserving its capacity to aggregate local and global spatio-temporal features, we design a parallel spike transformer block to replace sequential sub-blocks. Additionally, we propose a simplified spiking self-attention mechanism that omits the value parameter without compromising the model's performance. Experiments conducted on four datasets-PURE, UBFC-rPPG, UBFC-Phys, and MMPD demonstrate that the proposed model achieves a 12.4\% reduction in power consumption compared to PhysFormer. Additionally, the power consumption of the transformer block is reduced by a factor of 12.2, while maintaining decent performance as PhysFormer and other ANN-based models.
A Comprehensive Dataset and Automated Pipeline for Nailfold Capillary Analysis
Dec 10, 2023



Abstract:Nailfold capillaroscopy is a well-established method for assessing health conditions, but the untapped potential of automated medical image analysis using machine learning remains despite recent advancements. In this groundbreaking study, we present a pioneering effort in constructing a comprehensive dataset-321 images, 219 videos, 68 clinic reports, with expert annotations-that serves as a crucial resource for training deep-learning models. Leveraging this dataset, we propose an end-to-end nailfold capillary analysis pipeline capable of automatically detecting and measuring diverse morphological and dynamic features. Experimental results demonstrate sub-pixel measurement accuracy and 90% accuracy in predicting abnormality portions, highlighting its potential for advancing quantitative medical research and enabling pervasive computing in healthcare. We've shared our open-source codes and data (available at https://github.com/THU-CS-PI-LAB/ANFC-Automated-Nailfold-Capillary) to contribute to transformative progress in computational medical image analysis.
ALPHA: AnomaLous Physiological Health Assessment Using Large Language Models
Nov 21, 2023Abstract:This study concentrates on evaluating the efficacy of Large Language Models (LLMs) in healthcare, with a specific focus on their application in personal anomalous health monitoring. Our research primarily investigates the capabilities of LLMs in interpreting and analyzing physiological data obtained from FDA-approved devices. We conducted an extensive analysis using anomalous physiological data gathered in a simulated low-air-pressure plateau environment. This allowed us to assess the precision and reliability of LLMs in understanding and evaluating users' health status with notable specificity. Our findings reveal that LLMs exhibit exceptional performance in determining medical indicators, including a Mean Absolute Error (MAE) of less than 1 beat per minute for heart rate and less than 1% for oxygen saturation (SpO2). Furthermore, the Mean Absolute Percentage Error (MAPE) for these evaluations remained below 1%, with the overall accuracy of health assessments surpassing 85%. In image analysis tasks, such as interpreting photoplethysmography (PPG) data, our specially adapted GPT models demonstrated remarkable proficiency, achieving less than 1 bpm error in cycle count and 7.28 MAE for heart rate estimation. This study highlights LLMs' dual role as health data analysis tools and pivotal elements in advanced AI health assistants, offering personalized health insights and recommendations within the future health assistant framework.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge