Jeehyun Hwang
Time-IMM: A Dataset and Benchmark for Irregular Multimodal Multivariate Time Series
Jun 12, 2025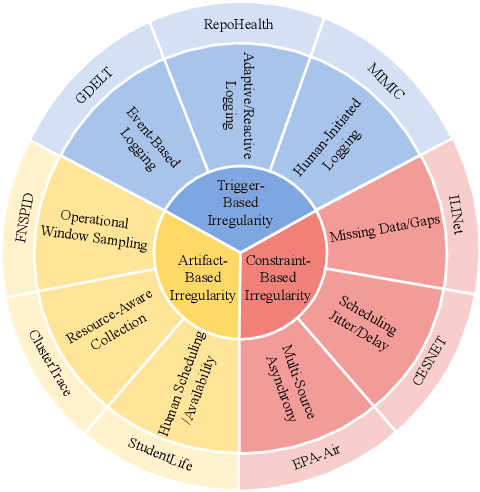
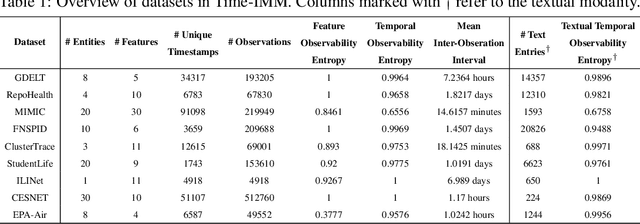
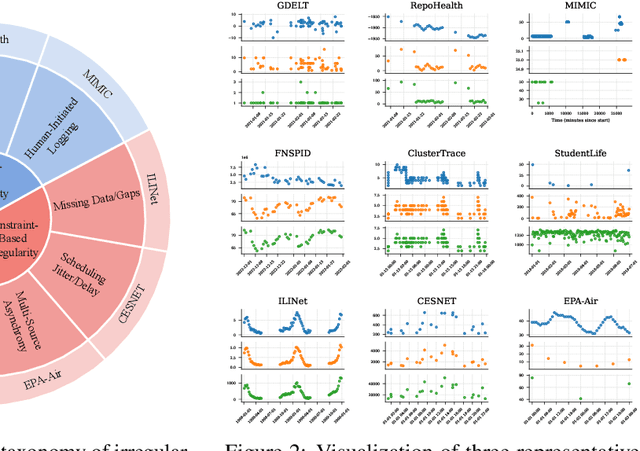
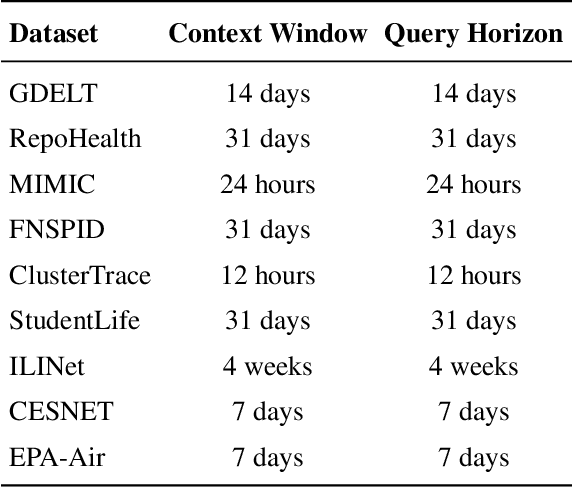
Abstract:Time series data in real-world applications such as healthcare, climate modeling, and finance are often irregular, multimodal, and messy, with varying sampling rates, asynchronous modalities, and pervasive missingness. However, existing benchmarks typically assume clean, regularly sampled, unimodal data, creating a significant gap between research and real-world deployment. We introduce Time-IMM, a dataset specifically designed to capture cause-driven irregularity in multimodal multivariate time series. Time-IMM represents nine distinct types of time series irregularity, categorized into trigger-based, constraint-based, and artifact-based mechanisms. Complementing the dataset, we introduce IMM-TSF, a benchmark library for forecasting on irregular multimodal time series, enabling asynchronous integration and realistic evaluation. IMM-TSF includes specialized fusion modules, including a timestamp-to-text fusion module and a multimodality fusion module, which support both recency-aware averaging and attention-based integration strategies. Empirical results demonstrate that explicitly modeling multimodality on irregular time series data leads to substantial gains in forecasting performance. Time-IMM and IMM-TSF provide a foundation for advancing time series analysis under real-world conditions. The dataset is publicly available at https://www.kaggle.com/datasets/blacksnail789521/time-imm/data, and the benchmark library can be accessed at https://anonymous.4open.science/r/IMMTSF_NeurIPS2025.
Causal Graph ODE: Continuous Treatment Effect Modeling in Multi-agent Dynamical Systems
Feb 29, 2024Abstract:Real-world multi-agent systems are often dynamic and continuous, where the agents co-evolve and undergo changes in their trajectories and interactions over time. For example, the COVID-19 transmission in the U.S. can be viewed as a multi-agent system, where states act as agents and daily population movements between them are interactions. Estimating the counterfactual outcomes in such systems enables accurate future predictions and effective decision-making, such as formulating COVID-19 policies. However, existing methods fail to model the continuous dynamic effects of treatments on the outcome, especially when multiple treatments (e.g., "stay-at-home" and "get-vaccine" policies) are applied simultaneously. To tackle this challenge, we propose Causal Graph Ordinary Differential Equations (CAG-ODE), a novel model that captures the continuous interaction among agents using a Graph Neural Network (GNN) as the ODE function. The key innovation of our model is to learn time-dependent representations of treatments and incorporate them into the ODE function, enabling precise predictions of potential outcomes. To mitigate confounding bias, we further propose two domain adversarial learning-based objectives, which enable our model to learn balanced continuous representations that are not affected by treatments or interference. Experiments on two datasets (i.e., COVID-19 and tumor growth) demonstrate the superior performance of our proposed model.
Learning Over Molecular Conformer Ensembles: Datasets and Benchmarks
Sep 29, 2023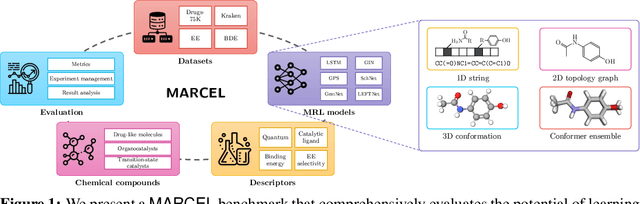
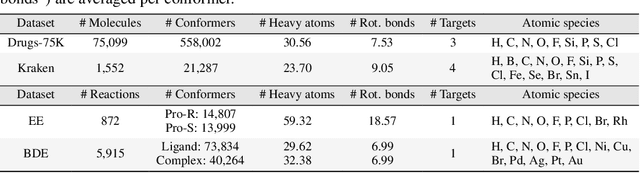
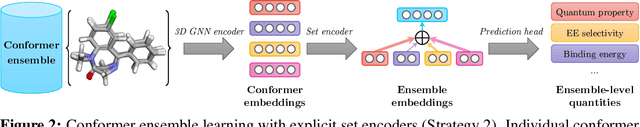
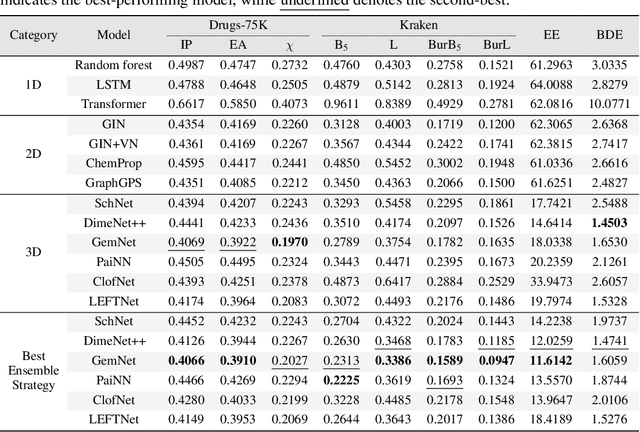
Abstract:Molecular Representation Learning (MRL) has proven impactful in numerous biochemical applications such as drug discovery and enzyme design. While Graph Neural Networks (GNNs) are effective at learning molecular representations from a 2D molecular graph or a single 3D structure, existing works often overlook the flexible nature of molecules, which continuously interconvert across conformations via chemical bond rotations and minor vibrational perturbations. To better account for molecular flexibility, some recent works formulate MRL as an ensemble learning problem, focusing on explicitly learning from a set of conformer structures. However, most of these studies have limited datasets, tasks, and models. In this work, we introduce the first MoleculAR Conformer Ensemble Learning (MARCEL) benchmark to thoroughly evaluate the potential of learning on conformer ensembles and suggest promising research directions. MARCEL includes four datasets covering diverse molecule- and reaction-level properties of chemically diverse molecules including organocatalysts and transition-metal catalysts, extending beyond the scope of common GNN benchmarks that are confined to drug-like molecules. In addition, we conduct a comprehensive empirical study, which benchmarks representative 1D, 2D, and 3D molecular representation learning models, along with two strategies that explicitly incorporate conformer ensembles into 3D MRL models. Our findings reveal that direct learning from an accessible conformer space can improve performance on a variety of tasks and models.
Graph Neural Controlled Differential Equations for Traffic Forecasting
Dec 07, 2021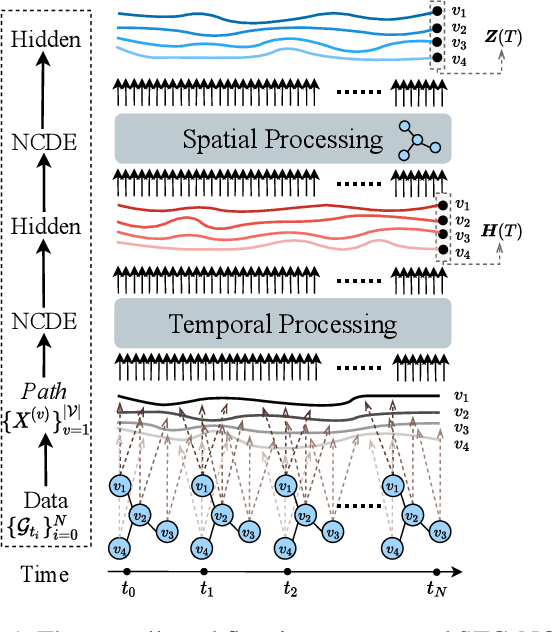
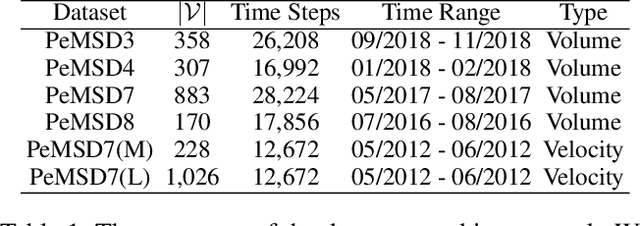
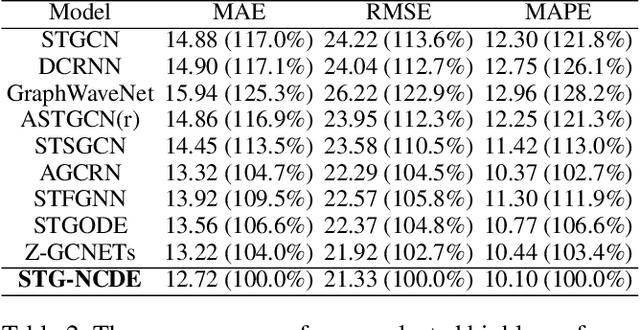
Abstract:Traffic forecasting is one of the most popular spatio-temporal tasks in the field of machine learning. A prevalent approach in the field is to combine graph convolutional networks and recurrent neural networks for the spatio-temporal processing. There has been fierce competition and many novel methods have been proposed. In this paper, we present the method of spatio-temporal graph neural controlled differential equation (STG-NCDE). Neural controlled differential equations (NCDEs) are a breakthrough concept for processing sequential data. We extend the concept and design two NCDEs: one for the temporal processing and the other for the spatial processing. After that, we combine them into a single framework. We conduct experiments with 6 benchmark datasets and 20 baselines. STG-NCDE shows the best accuracy in all cases, outperforming all those 20 baselines by non-trivial margins.
Climate Modeling with Neural Diffusion Equations
Nov 11, 2021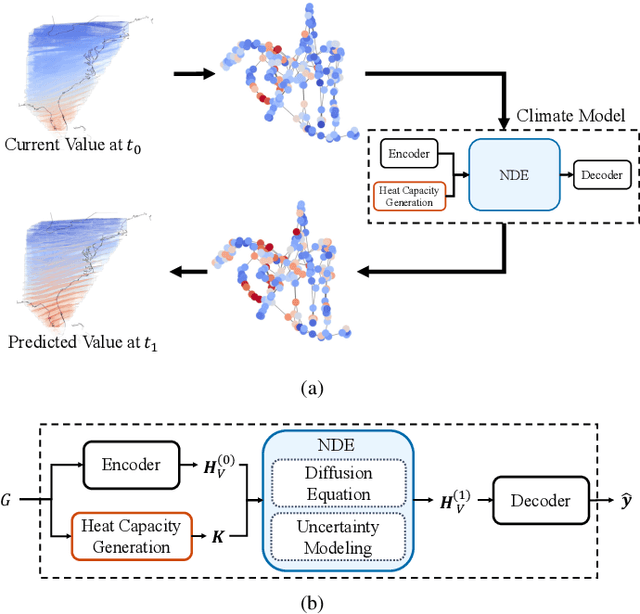

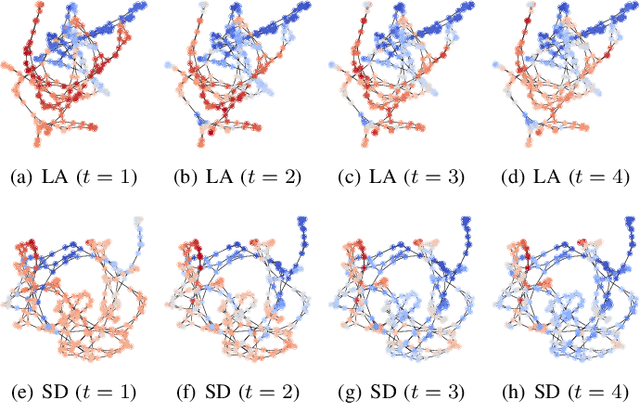
Abstract:Owing to the remarkable development of deep learning technology, there have been a series of efforts to build deep learning-based climate models. Whereas most of them utilize recurrent neural networks and/or graph neural networks, we design a novel climate model based on the two concepts, the neural ordinary differential equation (NODE) and the diffusion equation. Many physical processes involving a Brownian motion of particles can be described by the diffusion equation and as a result, it is widely used for modeling climate. On the other hand, neural ordinary differential equations (NODEs) are to learn a latent governing equation of ODE from data. In our presented method, we combine them into a single framework and propose a concept, called neural diffusion equation (NDE). Our NDE, equipped with the diffusion equation and one more additional neural network to model inherent uncertainty, can learn an appropriate latent governing equation that best describes a given climate dataset. In our experiments with two real-world and one synthetic datasets and eleven baselines, our method consistently outperforms existing baselines by non-trivial margins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge