Jae Won Choi
Advances in Automated Fetal Brain MRI Segmentation and Biometry: Insights from the FeTA 2024 Challenge
May 05, 2025



Abstract:Accurate fetal brain tissue segmentation and biometric analysis are essential for studying brain development in utero. The FeTA Challenge 2024 advanced automated fetal brain MRI analysis by introducing biometry prediction as a new task alongside tissue segmentation. For the first time, our diverse multi-centric test set included data from a new low-field (0.55T) MRI dataset. Evaluation metrics were also expanded to include the topology-specific Euler characteristic difference (ED). Sixteen teams submitted segmentation methods, most of which performed consistently across both high- and low-field scans. However, longitudinal trends indicate that segmentation accuracy may be reaching a plateau, with results now approaching inter-rater variability. The ED metric uncovered topological differences that were missed by conventional metrics, while the low-field dataset achieved the highest segmentation scores, highlighting the potential of affordable imaging systems when paired with high-quality reconstruction. Seven teams participated in the biometry task, but most methods failed to outperform a simple baseline that predicted measurements based solely on gestational age, underscoring the challenge of extracting reliable biometric estimates from image data alone. Domain shift analysis identified image quality as the most significant factor affecting model generalization, with super-resolution pipelines also playing a substantial role. Other factors, such as gestational age, pathology, and acquisition site, had smaller, though still measurable, effects. Overall, FeTA 2024 offers a comprehensive benchmark for multi-class segmentation and biometry estimation in fetal brain MRI, underscoring the need for data-centric approaches, improved topological evaluation, and greater dataset diversity to enable clinically robust and generalizable AI tools.
Unleashing the Strengths of Unlabeled Data in Pan-cancer Abdominal Organ Quantification: the FLARE22 Challenge
Aug 10, 2023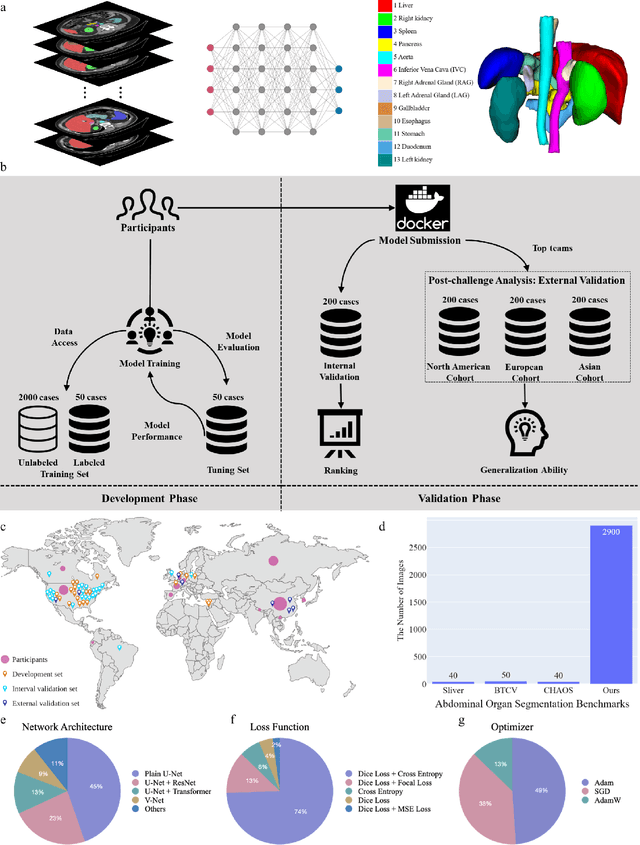
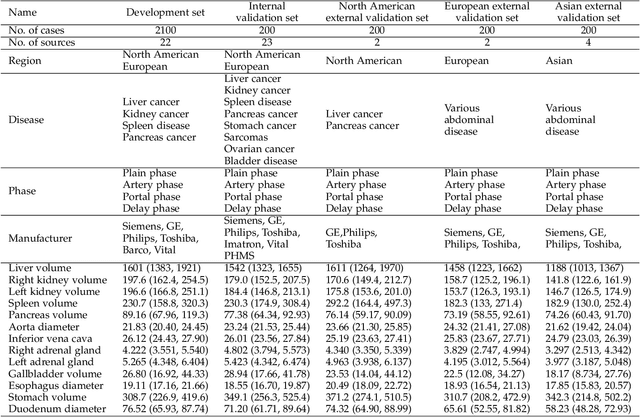
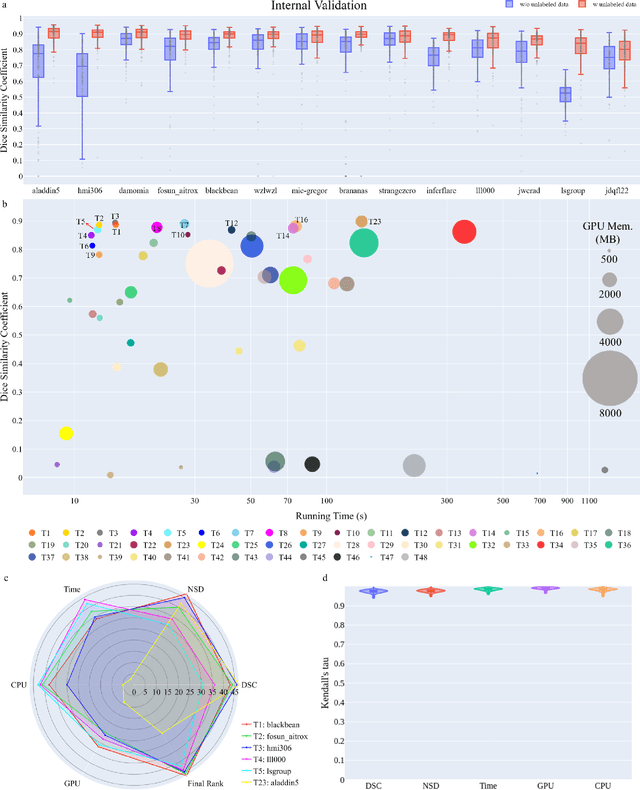
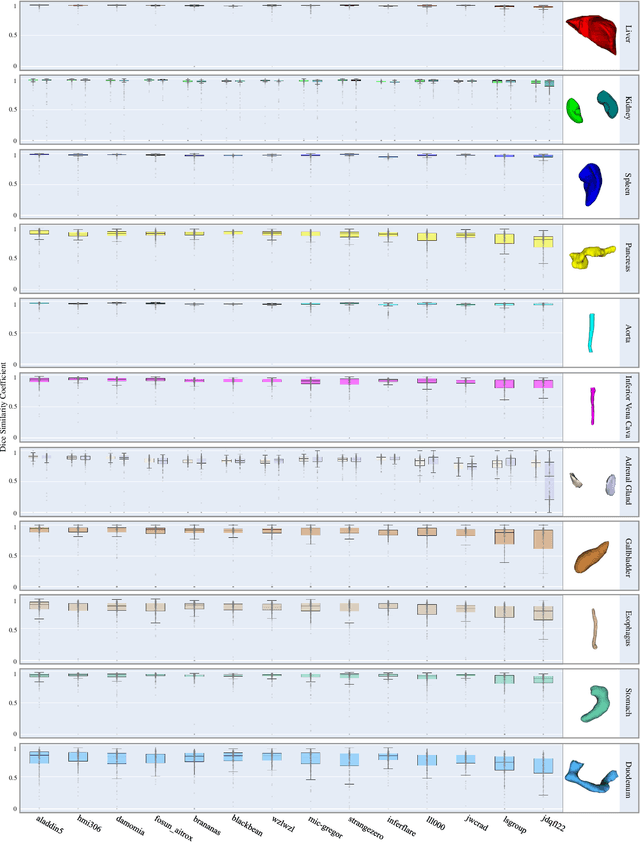
Abstract:Quantitative organ assessment is an essential step in automated abdominal disease diagnosis and treatment planning. Artificial intelligence (AI) has shown great potential to automatize this process. However, most existing AI algorithms rely on many expert annotations and lack a comprehensive evaluation of accuracy and efficiency in real-world multinational settings. To overcome these limitations, we organized the FLARE 2022 Challenge, the largest abdominal organ analysis challenge to date, to benchmark fast, low-resource, accurate, annotation-efficient, and generalized AI algorithms. We constructed an intercontinental and multinational dataset from more than 50 medical groups, including Computed Tomography (CT) scans with different races, diseases, phases, and manufacturers. We independently validated that a set of AI algorithms achieved a median Dice Similarity Coefficient (DSC) of 90.0\% by using 50 labeled scans and 2000 unlabeled scans, which can significantly reduce annotation requirements. The best-performing algorithms successfully generalized to holdout external validation sets, achieving a median DSC of 89.5\%, 90.9\%, and 88.3\% on North American, European, and Asian cohorts, respectively. They also enabled automatic extraction of key organ biology features, which was labor-intensive with traditional manual measurements. This opens the potential to use unlabeled data to boost performance and alleviate annotation shortages for modern AI models.
Online Segmented Recursive Least-Squares for Multipath Doppler Tracking
May 30, 2023


Abstract:Underwater communication signals typically suffer from distortion due to motion-induced Doppler. Especially in shallow water environments, recovering the signal is challenging due to the time-varying Doppler effects distorting each path differently. However, conventional Doppler estimation algorithms typically model uniform Doppler across all paths and often fail to provide robust Doppler tracking in multipath environments. In this paper, we propose a dynamic programming-inspired method, called online segmented recursive least-squares (OSRLS) to sequentially estimate the time-varying non-uniform Doppler across different multipath arrivals. By approximating the non-linear time distortion as a piece-wise-linear Markov model, we formulate the problem in a dynamic programming framework known as segmented least-squares (SLS). In order to circumvent an ill-conditioned formulation, perturbations are added to the Doppler model during the linearization process. The successful operation of the algorithm is demonstrated in a simulation on a synthetic channel with time-varying non-uniform Doppler.
Knowledge Distillation from Cross Teaching Teachers for Efficient Semi-Supervised Abdominal Organ Segmentation in CT
Nov 11, 2022Abstract:For more clinical applications of deep learning models for medical image segmentation, high demands on labeled data and computational resources must be addressed. This study proposes a coarse-to-fine framework with two teacher models and a student model that combines knowledge distillation and cross teaching, a consistency regularization based on pseudo-labels, for efficient semi-supervised learning. The proposed method is demonstrated on the abdominal multi-organ segmentation task in CT images under the MICCAI FLARE 2022 challenge, with mean Dice scores of 0.8429 and 0.8520 in the validation and test sets, respectively.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022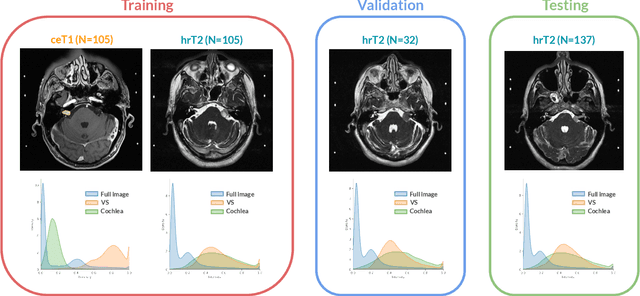
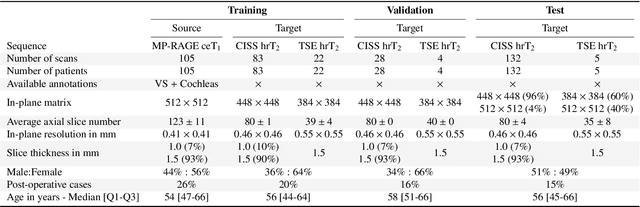
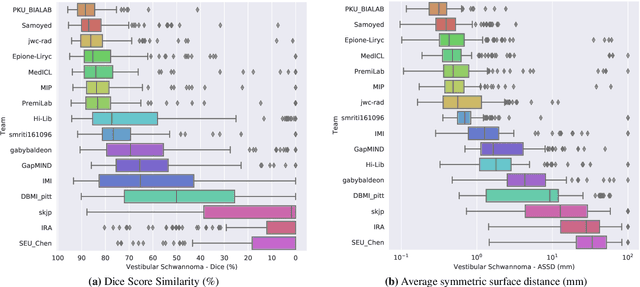
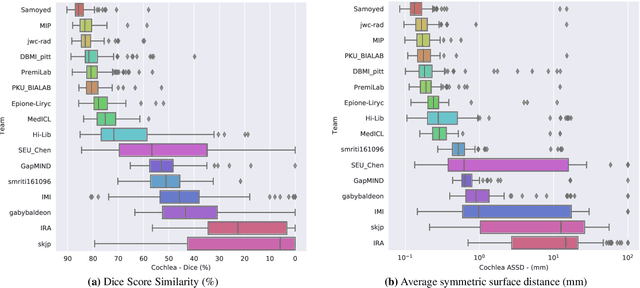
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
Using Out-of-the-Box Frameworks for Unpaired Image Translation and Image Segmentation for the crossMoDA Challenge
Oct 02, 2021
Abstract:The purpose of this study is to apply and evaluate out-of-the-box deep learning frameworks for the crossMoDA challenge. We use the CUT model for domain adaptation from contrast-enhanced T1 MR to high-resolution T2 MR. As data augmentation, we generated additional images with vestibular schwannomas with lower signal intensity. For the segmentation task, we use the nnU-Net framework. Our final submission achieved a mean Dice score of 0.8299 (0.0465) in the validation phase.
Video-Streaming Biomedical Implants using Ultrasonic Waves for Communication
Jun 28, 2021



Abstract:The use of wireless implanted medical devices (IMDs) is growing because they facilitate continuous monitoring of patients during normal activities, simplify medical procedures required for data retrieval and reduce the likelihood of infection associated with trailing wires. However, most of the state-of-the-art IMDs are passive and offline devices. One of the key obstacles to an active and online IMD is the infeasibility of real-time, high-quality video broadcast from the IMD. Such broadcast would help develop innovative devices such as a video-streaming capsule endoscopy (CE) pill with therapeutic intervention capabilities. State-of-the-art IMDs employ radio-frequency electromagnetic waves for information transmission. However, high attenuation of RF-EM waves in tissues and federal restrictions on the transmit power and operable bandwidth lead to fundamental performance constraints for IMDs employing RF links, and prevent achieving high data rates that could accomodate video broadcast. In this work, ultrasonic waves were used for video transmission and broadcast through biological tissues. The proposed proof-of-concept system was tested on a porcine intestine ex vivo and a rabbit in vivo. It was demonstrated that using a millimeter-sized, implanted biocompatible transducer operating at 1.1-1.2 MHz, it was possible to transmit endoscopic video with high resolution (1280 pixels by 720 pixels) through porcine intestine wrapped with bacon, and to broadcast standard definition (640 pixels by 480 pixels) video near real-time through rabbit abdomen in vivo. A media repository that includes experimental demonstrations and media files accompanies this paper. The accompanying media repository can be found at this link: https://bit.ly/3wuc7tk.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge