Michael L. Oelze
Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana IL, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana IL, Carle Illinois College of Medicine, University of Illinois at Urbana-Champaign, Urbana IL
Label-free Super-Resolution Microvessel Color Flow Imaging with Ultrasound
May 27, 2025

Abstract:We present phase subtraction imaging (PSI), a new spatial-temporal beamforming method that enables micrometer level resolution imaging of microvessels in live animals without labels, which are microbubbles in ultrasound super-resolution imaging. Subtraction of relative phase differences between consecutive frames beamformed with mismatched apodizations is used in PSI to overcome the diffraction limit. We validated this method by imaging both the mouse brain and rabbit kidney using different ultrasound probes and scanning machines.
The Art of the Steal: Purloining Deep Learning Models Developed for an Ultrasound Scanner to a Competitor Machine
Jul 03, 2024Abstract:A transfer function approach has recently proven effective for calibrating deep learning (DL) algorithms in quantitative ultrasound (QUS), addressing data shifts at both the acquisition and machine levels. Expanding on this approach, we develop a strategy to 'steal' the functionality of a DL model from one ultrasound machine and implement it on another, in the context of QUS. This demonstrates the ease with which the functionality of a DL model can be transferred between machines, highlighting the security risks associated with deploying such models in a commercial scanner for clinical use. The proposed method is a black-box unsupervised domain adaptation technique that integrates the transfer function approach with an iterative schema. It does not utilize any information related to model internals of the victim machine but it solely relies on the availability of input-output interface. Additionally, we assume the availability of unlabelled data from the testing machine, i.e., the perpetrator machine. This scenario could become commonplace as companies begin deploying their DL functionalities for clinical use. Competing companies might acquire the victim machine and, through the input-output interface, replicate the functionality onto their own machines. In the experiments, we used a SonixOne and a Verasonics machine. The victim model was trained on SonixOne data, and its functionality was then transferred to the Verasonics machine. The proposed method successfully transferred the functionality to the Verasonics machine, achieving a remarkable 98\% classification accuracy in a binary decision task. This study underscores the need to establish security measures prior to deploying DL models in clinical settings.
Machine-to-Machine Transfer Function in Deep Learning-Based Quantitative Ultrasound
Nov 27, 2023Abstract:A Transfer Function approach was recently demonstrated to mitigate data mismatches at the acquisition level for a single ultrasound scanner in deep learning (DL) based quantitative ultrasound (QUS). As a natural progression, we further investigate the transfer function approach and introduce a Machine-to-Machine (M2M) Transfer Function, which possesses the ability to mitigate data mismatches at a machine level, i.e., mismatches between two scanners over the same frequency band. This ability opens the door to unprecedented opportunities for reducing DL model development costs, enabling the combination of data from multiple sources or scanners, or facilitating the transfer of DL models between machines with ease. We tested the proposed method utilizing a SonixOne machine and a Verasonics machine. In the experiments, we used a L9-4 array and conducted two types of acquisitions to obtain calibration data: stable and free-hand, using two different calibration phantoms. Without the proposed calibration method, the mean classification accuracy when applying a model on data acquired from one system to data acquired from another system was approximately 50%, and the mean AUC was about 0.40. With the proposed method, mean accuracy increased to approximately 90%, and the AUC rose to the 0.99. Additional observations include that shifts in statistics for the z-score normalization had a significant impact on performance. Furthermore, the choice of the calibration phantom played an important role in the proposed method. Additionally, robust implementation inspired by Wiener filtering provided an effective method for transferring the domain from one machine to another machine, and it can succeed using just a single calibration view without the need for multiple independent calibration frames.
A Radiological Clip Design Using Ultrasound Identification to Improve Localization
Aug 30, 2023



Abstract:Objective: We demonstrate the use of ultrasound to receive an acoustic signal transmitted from a radiological clip designed from a custom circuit. This signal encodes an identification number and is localized and identified wirelessly by the ultrasound imaging system. Methods: We designed and constructed the test platform with a Teensy 4.0 microcontroller core to detect ultrasonic imaging pulses received by a transducer embedded in a phantom, which acted as the radiological clip. Ultrasound identification (USID) signals were generated and transmitted as a result. The phantom and clip were imaged using an ultrasonic array (Philips L7-4) connected to a Verasonics Vantage 128 system operating in pulse inversion (PI) mode. Cross-correlations were performed to localize and identify the code sequences in the PI images. Results: USID signals were detected and visualized on B-mode images of the phantoms with up to sub-millimeter localization accuracy. The average detection rate across 1,600 frames of ultrasound data was 94.6%. Tested ID values exhibited differences in detection rates. Conclusion: The USID clip produced identifiable, distinguishable, and localizable signals when imaged. Significance: Radiological clips are used to mark breast cancer being treated by neoadjuvant chemotherapy (NAC) via implant in or near treated lesions. As NAC progresses, available marking clips can lose visibility in ultrasound, the imaging modality of choice for monitoring NAC-treated lesions. By transmitting an active signal, more accurate and reliable ultrasound localization of these clips could be achieved and multiple clips with different ID values could be imaged in the same field of view.
High-resolution Power Doppler Using Null Subtraction Imaging
Jan 09, 2023



Abstract:To improve the spatial resolution of power Doppler (PD) imaging, we explored null subtraction imaging (NSI) as an alternative beamforming technique to delay-and-sum (DAS). NSI is a nonlinear beamforming approach that uses three different apodizations on receive and incoherently sums the beamformed envelopes. NSI uses a null in the beam pattern to improve the lateral resolution, which we apply here for improving PD spatial resolution both with and without contrast microbubbles. In this study, we used NSI with singular value decomposition (SVD)-based clutter filtering and noise equalization to generate high-resolution PD images. An element sensitivity correction scheme was also performed to further improve the image quality of PD images using NSI. First, a microbubble trace experiment was performed to quantitatively evaluate the performance of NSI based PD. Then, both contrast-enhanced and contrast free ultrasound data were collected from a rat brain. Higher spatial resolution and image quality were observed from the NSI-based PD microvessel images compared to microvessel images generated by traditional DAS-based beamforming.
Grating lobe reduction in plane wave imaging with angular compounding using subtraction of coherent signals
Oct 24, 2022

Abstract:Plane wave imaging (PWI) with angular compounding has gained in popularity over recent years because it provides high frame rates and good image properties. However, most linear arrays used in clinical practice have a pitch that is equal to than the wavelength of ultrasound. Hence, the presence of grating lobes is a concern for PWI using multiple transmit angles. The presence of grating lobes produces clutter in images and reduces the ability to observe tissue contrast. Techniques to reduce or eliminate the presence of grating lobes for PWI using multiple angles will result in improved image quality. Null subtraction imaging (NSI) is a nonlinear beamforming technique that has been explored for improving the lateral resolution of ultrasonic imaging. However, the apodization scheme used in NSI also eliminates or greatly reduces the presence of grating lobes. Imaging tasks using NSI were evaluated in simulations and physical experiments involving tissue-mimicking phantoms and rat tumors in vivo. Images created with NSI were compared with images created using traditional delay and sum (DAS) with Hann apodization and images created using a generalized coherence factor (GCF). NSI was observed to greatly reduce the presence of grating lobes in ultrasonic images, compared to DAS with Hann and GCF, while maintaining spatial resolution and contrast in the images. Therefore, NSI can provide a novel means of creating images using PWI with multiple steering angles on clinically available linear arrays while reducing the adverse effects associated with grating lobes.
Through Tissue Ultra-high-definition Video Transmission Using an Ultrasound Communication Channel
Oct 18, 2022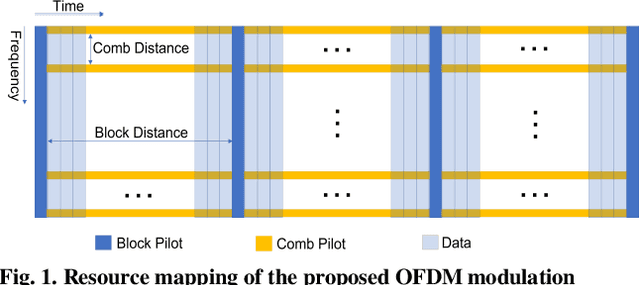

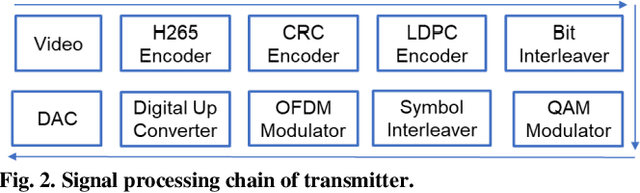
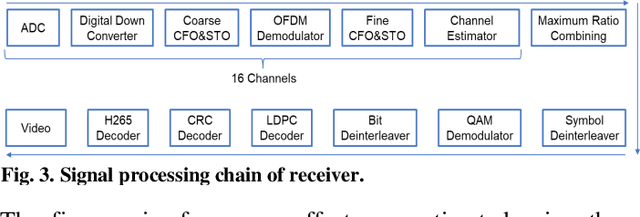
Abstract:Wireless capsule endoscopy (WCE) has been widely adopted as complementary to traditional wired gastroendoscopy, especially for small bowel diseases which are beyond the latter's reach. However, both the video resolution and frame rates are limited in current WCE solutions due to the limited wireless data rate. The reasons behind this are that the electromagnetic (EM), radio frequency (RF) based communication scheme used by WCE has strict limits on useable bandwidth and power, and the high attenuation in the human body compared to air. Ultrasound communication could be a potential alternative solution as it has access to much higher bandwidths and transmitted power with much lower attenuation. In this paper, we propose an ultrasound communication scheme specially designed for high data rate through tissue data transmission and validate this communication scheme by successfully transmitting ultra-high-definition (UHD) video (3840*2160 pixels at 60 FPS) through 5 cm of pork belly. Over 8.3 Mbps error free payload data rate was achieved with the proposed communication scheme and our custom-built field programmable gate array (FPGA) based test platform.
Calibrating Data Mismatches in Deep Learning-Based Quantitative Ultrasound Using Setting Transfer Functions
Oct 04, 2022
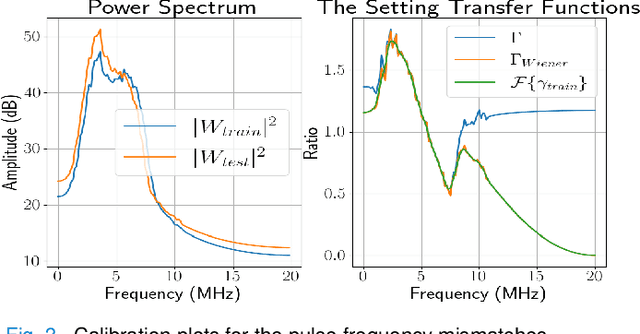
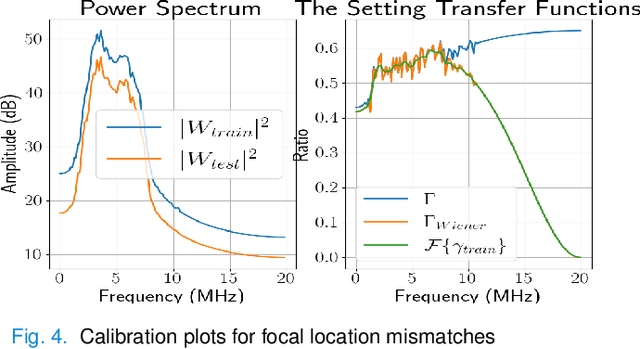
Abstract:Deep learning (DL) can fail when there are data mismatches between training and testing data. Due to its operator-dependent nature, acquisition-related data mismatches, caused by different scanner settings, can occur in ultrasound imaging. Therefore, mitigating effects of such data mismatches is essential for wider clinical adoption of DL powered ultrasound imaging. To mitigate the effects, ideally we need to collect a large training set at each scanner setting. However, acquiring such training sets is expensive. Another approach could be training on a subset of imaging settings, which makes the data generation less expensive. However, there will still be generalization issues. As an alternative approach that is inexpensive and generalizable, we propose to collect a large training set at a single setting and a small calibration set at each scanner setting. Then, the calibration set will be used to calibrate data mismatches by using a signals and systems perspective. We tested the proposed solution to classify two phantoms. To investigate generalizability of the proposed solution, we calibrated three types of data mismatches: pulse frequency, focus and output power mismatches. To calibrate the setting mismatches, we calculated the setting transfer functions. The CNN trained with no calibration resulted in mean classification accuracies of 55.3%, 64.4% and 70.3% for pulse frequency, focus and output power mismatches, respectively. By using the setting transfer functions, which allowed a matching of the training and testing domains, we obtained mean accuracies of 95.3%, 92.99% and 99.32%, respectively. Therefore, the incorporation of the setting transfer functions between scanner settings can provide an economical means of generalizing a DL model for specific classification tasks where scanner settings are not fixed by the operator.
Towards a real-time continuous ultrafast ultrasound beamformer with programmable logic
Aug 06, 2022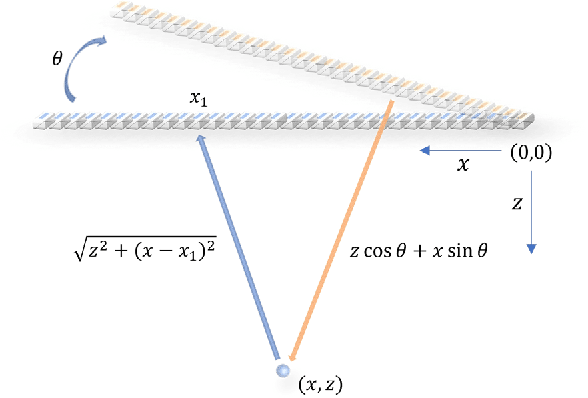
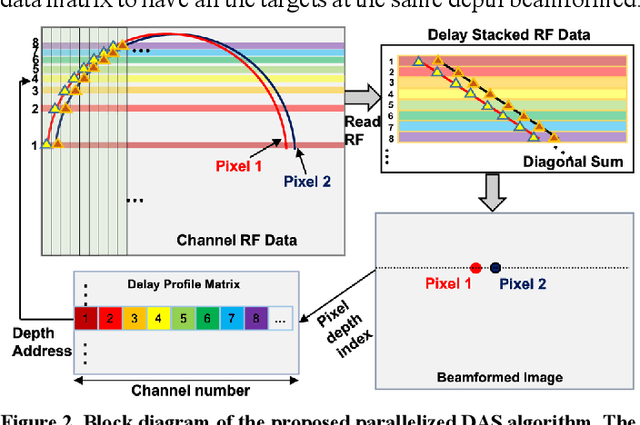
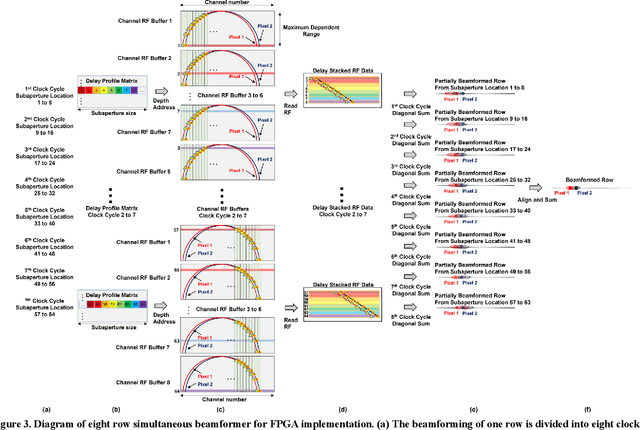
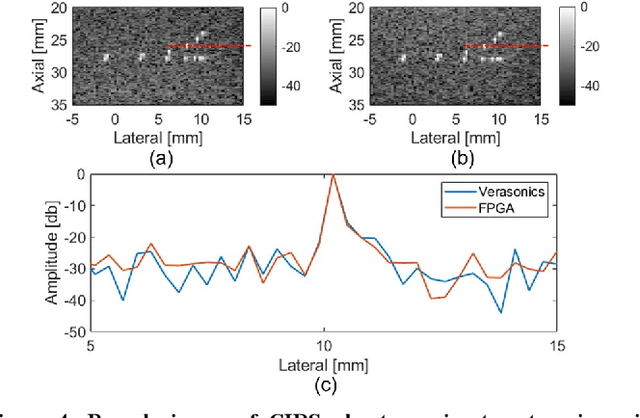
Abstract:Ultrafast ultrasound imaging is essential for advanced ultrasound imaging techniques such as ultrasound localization microscopy (ULM) and functional ultrasound (fUS). Current ultrafast ultrasound imaging is challenged by the ultrahigh data bandwidth associated with the radio frequency (RF) signal, and by the latency of the computationally expensive beamforming process. As such, continuous ultrafast data acquisition and beamforming remain elusive with existing software beamformers based on CPUs or GPUs. To address these challenges, the proposed work introduces a hybrid solution composed of an improved delay and sum (DAS) algorithm with high hardware efficiency and an ultrafast beamformer based on the field programmable gate array (FPGA). Our proposed method presents two unique advantages over conventional FPGA-based beamformers: 1) high scalability that allows fast adaptation to different FPGA platforms; 2) high adaptability to different imaging probes and applications thanks to the absence of hard-coded imaging parameters. With the proposed method, we measured an ultrafast beamforming frame rate of over 3.38 GPixels/second. The performance of the proposed beamformer was compared with the software beamformer on the Verasonics Vantage system for both phantom imaging and in vivo imaging of a mouse brain. Multiple imaging schemes including B-mode, power Doppler and ULM were evaluated with the proposed solution.
A Data-Efficient Deep Learning Training Strategy for Biomedical Ultrasound Imaging: Zone Training
Feb 01, 2022



Abstract:Deep learning (DL) powered biomedical ultrasound imaging is an emerging research field where researchers adapt the image analysis capabilities of DL algorithms to biomedical ultrasound imaging settings. A major roadblock to wider adoption of DL powered biomedical ultrasound imaging is that acquiring large and diverse datasets is expensive in clinical settings, which is a requirement for successful DL implementation. Hence, there is a constant need for developing data-efficient DL techniques to turn DL powered biomedical ultrasound imaging into reality. In this work, we develop a data-efficient deep learning training strategy, which we named \textit{Zone Training}. In \textit{Zone Training}, we propose to divide the complete field of view of an ultrasound image into multiple zones associated with different regions of a diffraction pattern and then, train separate DL networks for each zone. The main advantage of \textit{Zone Training} is that it requires less training data to achieve high accuracy. In this work, three different tissue-mimicking phantoms were classified by a DL network. The results demonstrated that \textit{Zone Training} required a factor of 2-5 less training data to achieve similar classification accuracies compared to a conventional training strategy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge