Isabella Nogues
3D Graph Anatomy Geometry-Integrated Network for Pancreatic Mass Segmentation, Diagnosis, and Quantitative Patient Management
Dec 08, 2020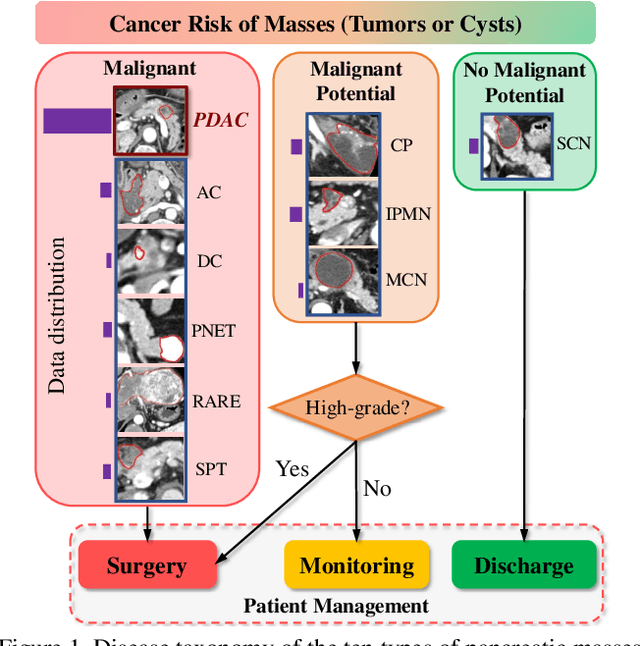
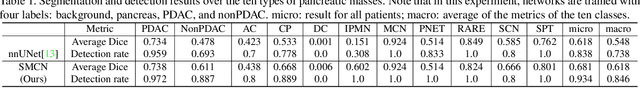
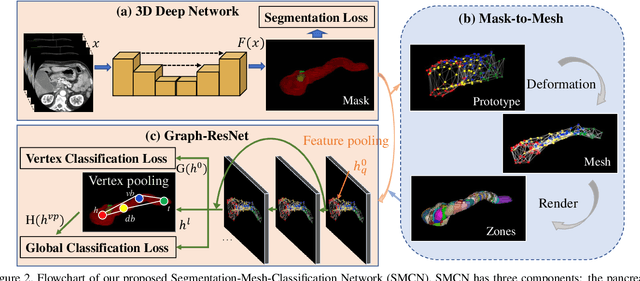
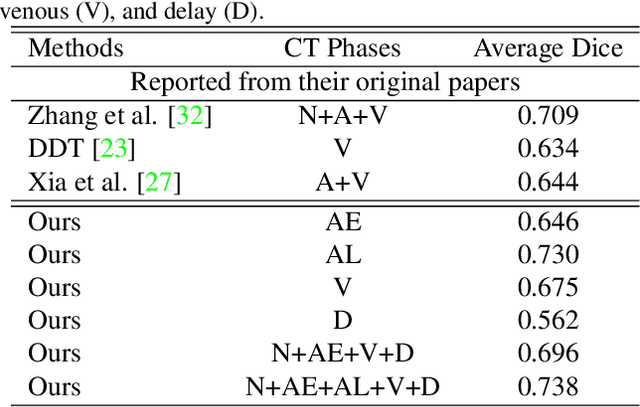
Abstract:The pancreatic disease taxonomy includes ten types of masses (tumors or cysts)[20,8]. Previous work focuses on developing segmentation or classification methods only for certain mass types. Differential diagnosis of all mass types is clinically highly desirable [20] but has not been investigated using an automated image understanding approach. We exploit the feasibility to distinguish pancreatic ductal adenocarcinoma (PDAC) from the nine other nonPDAC masses using multi-phase CT imaging. Both image appearance and the 3D organ-mass geometry relationship are critical. We propose a holistic segmentation-mesh-classification network (SMCN) to provide patient-level diagnosis, by fully utilizing the geometry and location information, which is accomplished by combining the anatomical structure and the semantic detection-by-segmentation network. SMCN learns the pancreas and mass segmentation task and builds an anatomical correspondence-aware organ mesh model by progressively deforming a pancreas prototype on the raw segmentation mask (i.e., mask-to-mesh). A new graph-based residual convolutional network (Graph-ResNet), whose nodes fuse the information of the mesh model and feature vectors extracted from the segmentation network, is developed to produce the patient-level differential classification results. Extensive experiments on 661 patients' CT scans (five phases per patient) show that SMCN can improve the mass segmentation and detection accuracy compared to the strong baseline method nnUNet (e.g., for nonPDAC, Dice: 0.611 vs. 0.478; detection rate: 89% vs. 70%), achieve similar sensitivity and specificity in differentiating PDAC and nonPDAC as expert radiologists (i.e., 94% and 90%), and obtain results comparable to a multimodality test [20] that combines clinical, imaging, and molecular testing for clinical management of patients.
DeepPap: Deep Convolutional Networks for Cervical Cell Classification
Jan 25, 2018



Abstract:Automation-assisted cervical screening via Pap smear or liquid-based cytology (LBC) is a highly effective cell imaging based cancer detection tool, where cells are partitioned into "abnormal" and "normal" categories. However, the success of most traditional classification methods relies on the presence of accurate cell segmentations. Despite sixty years of research in this field, accurate segmentation remains a challenge in the presence of cell clusters and pathologies. Moreover, previous classification methods are only built upon the extraction of hand-crafted features, such as morphology and texture. This paper addresses these limitations by proposing a method to directly classify cervical cells - without prior segmentation - based on deep features, using convolutional neural networks (ConvNets). First, the ConvNet is pre-trained on a natural image dataset. It is subsequently fine-tuned on a cervical cell dataset consisting of adaptively re-sampled image patches coarsely centered on the nuclei. In the testing phase, aggregation is used to average the prediction scores of a similar set of image patches. The proposed method is evaluated on both Pap smear and LBC datasets. Results show that our method outperforms previous algorithms in classification accuracy (98.3%), area under the curve (AUC) (0.99) values, and especially specificity (98.3%), when applied to the Herlev benchmark Pap smear dataset and evaluated using five-fold cross-validation. Similar superior performances are also achieved on the HEMLBC (H&E stained manual LBC) dataset. Our method is promising for the development of automation-assisted reading systems in primary cervical screening.
Unsupervised Joint Mining of Deep Features and Image Labels for Large-scale Radiology Image Categorization and Scene Recognition
Dec 27, 2017
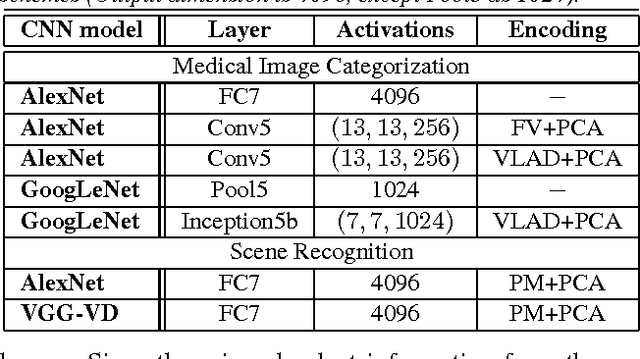
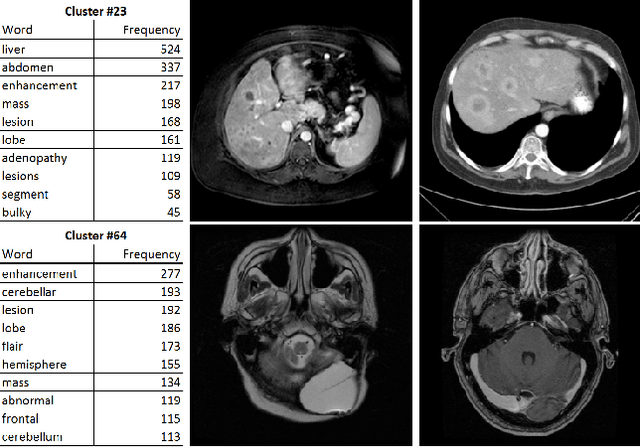
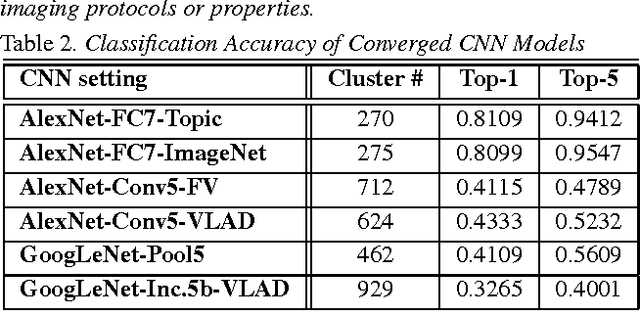
Abstract:The recent rapid and tremendous success of deep convolutional neural networks (CNN) on many challenging computer vision tasks largely derives from the accessibility of the well-annotated ImageNet and PASCAL VOC datasets. Nevertheless, unsupervised image categorization (i.e., without the ground-truth labeling) is much less investigated, yet critically important and difficult when annotations are extremely hard to obtain in the conventional way of "Google Search" and crowd sourcing. We address this problem by presenting a looped deep pseudo-task optimization (LDPO) framework for joint mining of deep CNN features and image labels. Our method is conceptually simple and rests upon the hypothesized "convergence" of better labels leading to better trained CNN models which in turn feed more discriminative image representations to facilitate more meaningful clusters/labels. Our proposed method is validated in tackling two important applications: 1) Large-scale medical image annotation has always been a prohibitively expensive and easily-biased task even for well-trained radiologists. Significantly better image categorization results are achieved via our proposed approach compared to the previous state-of-the-art method. 2) Unsupervised scene recognition on representative and publicly available datasets with our proposed technique is examined. The LDPO achieves excellent quantitative scene classification results. On the MIT indoor scene dataset, it attains a clustering accuracy of 75.3%, compared to the state-of-the-art supervised classification accuracy of 81.0% (when both are based on the VGG-VD model).
Unsupervised Category Discovery via Looped Deep Pseudo-Task Optimization Using a Large Scale Radiology Image Database
Mar 25, 2016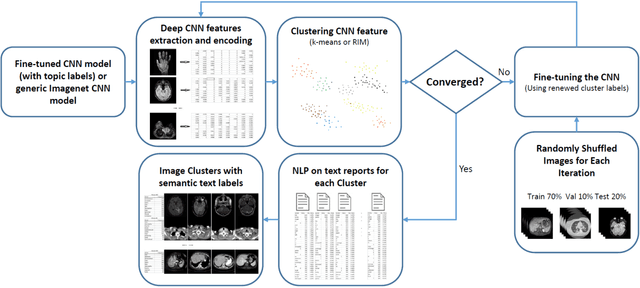
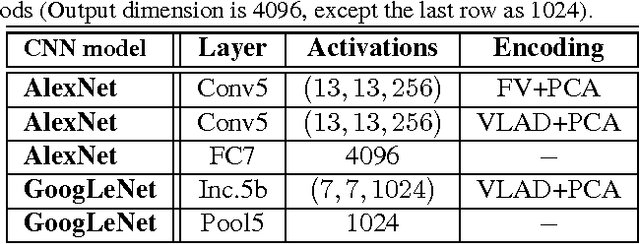
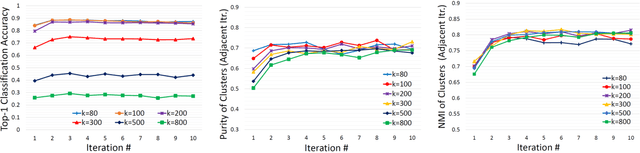
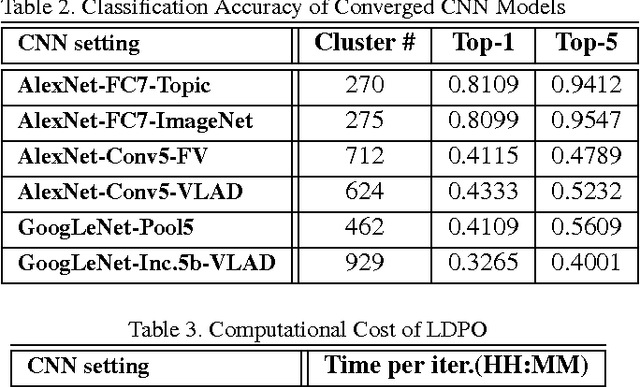
Abstract:Obtaining semantic labels on a large scale radiology image database (215,786 key images from 61,845 unique patients) is a prerequisite yet bottleneck to train highly effective deep convolutional neural network (CNN) models for image recognition. Nevertheless, conventional methods for collecting image labels (e.g., Google search followed by crowd-sourcing) are not applicable due to the formidable difficulties of medical annotation tasks for those who are not clinically trained. This type of image labeling task remains non-trivial even for radiologists due to uncertainty and possible drastic inter-observer variation or inconsistency. In this paper, we present a looped deep pseudo-task optimization procedure for automatic category discovery of visually coherent and clinically semantic (concept) clusters. Our system can be initialized by domain-specific (CNN trained on radiology images and text report derived labels) or generic (ImageNet based) CNN models. Afterwards, a sequence of pseudo-tasks are exploited by the looped deep image feature clustering (to refine image labels) and deep CNN training/classification using new labels (to obtain more task representative deep features). Our method is conceptually simple and based on the hypothesized "convergence" of better labels leading to better trained CNN models which in turn feed more effective deep image features to facilitate more meaningful clustering/labels. We have empirically validated the convergence and demonstrated promising quantitative and qualitative results. Category labels of significantly higher quality than those in previous work are discovered. This allows for further investigation of the hierarchical semantic nature of the given large-scale radiology image database.
Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning
Feb 10, 2016
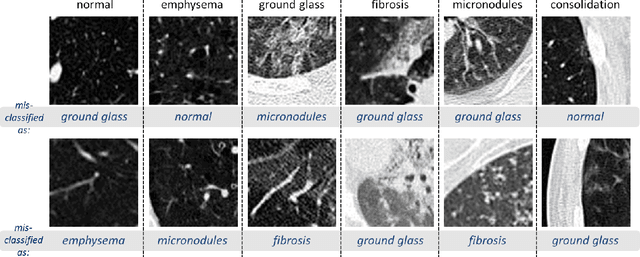

Abstract:Remarkable progress has been made in image recognition, primarily due to the availability of large-scale annotated datasets and the revival of deep CNN. CNNs enable learning data-driven, highly representative, layered hierarchical image features from sufficient training data. However, obtaining datasets as comprehensively annotated as ImageNet in the medical imaging domain remains a challenge. There are currently three major techniques that successfully employ CNNs to medical image classification: training the CNN from scratch, using off-the-shelf pre-trained CNN features, and conducting unsupervised CNN pre-training with supervised fine-tuning. Another effective method is transfer learning, i.e., fine-tuning CNN models pre-trained from natural image dataset to medical image tasks. In this paper, we exploit three important, but previously understudied factors of employing deep convolutional neural networks to computer-aided detection problems. We first explore and evaluate different CNN architectures. The studied models contain 5 thousand to 160 million parameters, and vary in numbers of layers. We then evaluate the influence of dataset scale and spatial image context on performance. Finally, we examine when and why transfer learning from pre-trained ImageNet (via fine-tuning) can be useful. We study two specific computer-aided detection (CADe) problems, namely thoraco-abdominal lymph node (LN) detection and interstitial lung disease (ILD) classification. We achieve the state-of-the-art performance on the mediastinal LN detection, with 85% sensitivity at 3 false positive per patient, and report the first five-fold cross-validation classification results on predicting axial CT slices with ILD categories. Our extensive empirical evaluation, CNN model analysis and valuable insights can be extended to the design of high performance CAD systems for other medical imaging tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge