Shaoxiong Liu
Reversed Active Learning based Atrous DenseNet for Pathological Image Classification
Jul 06, 2018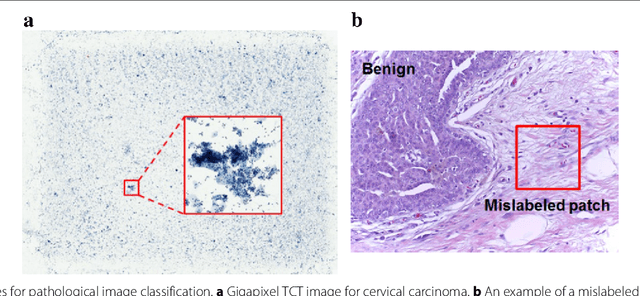
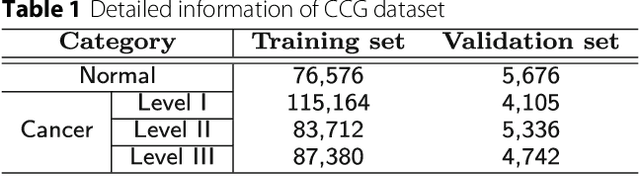
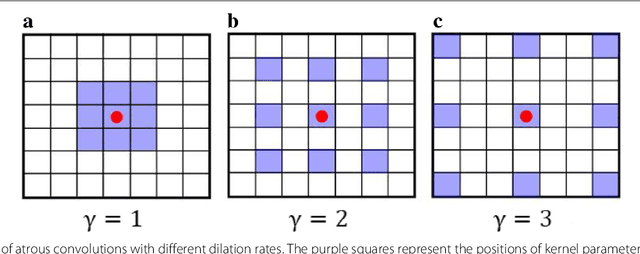
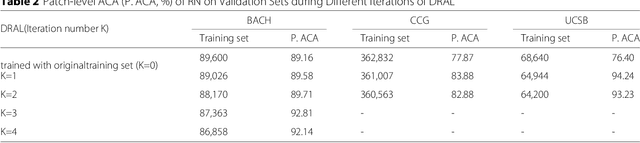
Abstract:Witnessed the development of deep learning in recent years, increasing number of researches try to adopt deep learning model for medical image analysis. However, the usage of deep learning networks for the pathological image analysis encounters several challenges, e.g. high resolution (gigapixel) of pathological images and lack of annotations of cancer areas. To address the challenges, we proposed a complete framework for the pathological image classification, which consists of a novel training strategy, namely reversed active learning (RAL), and an advanced network, namely atrous DenseNet (ADN). The proposed RAL can remove the mislabel patches in the training set. The refined training set can then be used to train widely used deep learning networks, e.g. VGG-16, ResNets, etc. A novel deep learning network, i.e. atrous DenseNet (ADN), is also proposed for the classification of pathological images. The proposed ADN achieves multi-scale feature extraction by integrating the atrous convolutions to the Dense Block. The proposed RAL and ADN have been evaluated on two pathological datasets, i.e. BACH and CCG. The experimental results demonstrate the excellent performance of the proposed ADN + RAL framework, i.e. the average patch-level ACAs of 94.10% and 92.05% on BACH and CCG validation sets were achieved.
DeepPap: Deep Convolutional Networks for Cervical Cell Classification
Jan 25, 2018



Abstract:Automation-assisted cervical screening via Pap smear or liquid-based cytology (LBC) is a highly effective cell imaging based cancer detection tool, where cells are partitioned into "abnormal" and "normal" categories. However, the success of most traditional classification methods relies on the presence of accurate cell segmentations. Despite sixty years of research in this field, accurate segmentation remains a challenge in the presence of cell clusters and pathologies. Moreover, previous classification methods are only built upon the extraction of hand-crafted features, such as morphology and texture. This paper addresses these limitations by proposing a method to directly classify cervical cells - without prior segmentation - based on deep features, using convolutional neural networks (ConvNets). First, the ConvNet is pre-trained on a natural image dataset. It is subsequently fine-tuned on a cervical cell dataset consisting of adaptively re-sampled image patches coarsely centered on the nuclei. In the testing phase, aggregation is used to average the prediction scores of a similar set of image patches. The proposed method is evaluated on both Pap smear and LBC datasets. Results show that our method outperforms previous algorithms in classification accuracy (98.3%), area under the curve (AUC) (0.99) values, and especially specificity (98.3%), when applied to the Herlev benchmark Pap smear dataset and evaluated using five-fold cross-validation. Similar superior performances are also achieved on the HEMLBC (H&E stained manual LBC) dataset. Our method is promising for the development of automation-assisted reading systems in primary cervical screening.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge