Ronald Summers
Enhancing chest X-ray datasets with privacy-preserving large language models and multi-type annotations: a data-driven approach for improved classification
Mar 06, 2024Abstract:In chest X-ray (CXR) image analysis, rule-based systems are usually employed to extract labels from reports, but concerns exist about label quality. These datasets typically offer only presence labels, sometimes with binary uncertainty indicators, which limits their usefulness. In this work, we present MAPLEZ (Medical report Annotations with Privacy-preserving Large language model using Expeditious Zero shot answers), a novel approach leveraging a locally executable Large Language Model (LLM) to extract and enhance findings labels on CXR reports. MAPLEZ extracts not only binary labels indicating the presence or absence of a finding but also the location, severity, and radiologists' uncertainty about the finding. Over eight abnormalities from five test sets, we show that our method can extract these annotations with an increase of 5 percentage points (pp) in F1 score for categorical presence annotations and more than 30 pp increase in F1 score for the location annotations over competing labelers. Additionally, using these improved annotations in classification supervision, we demonstrate substantial advancements in model quality, with an increase of 1.7 pp in AUROC over models trained with annotations from the state-of-the-art approach. We share code and annotations.
RATCHET: Medical Transformer for Chest X-ray Diagnosis and Reporting
Jul 05, 2021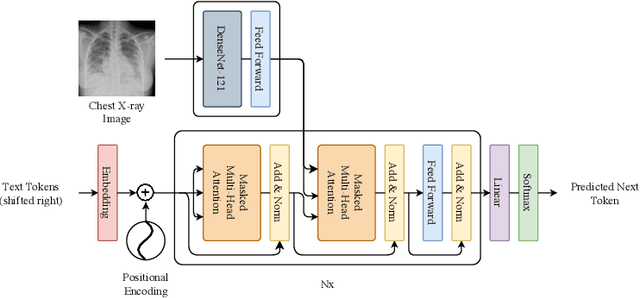
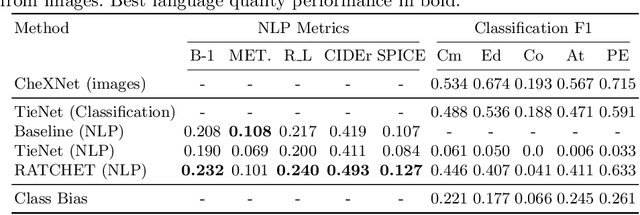
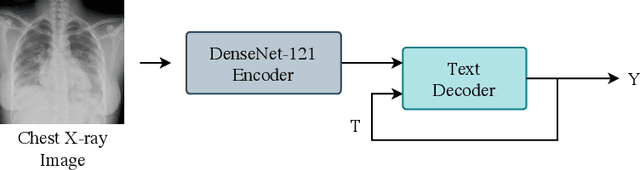
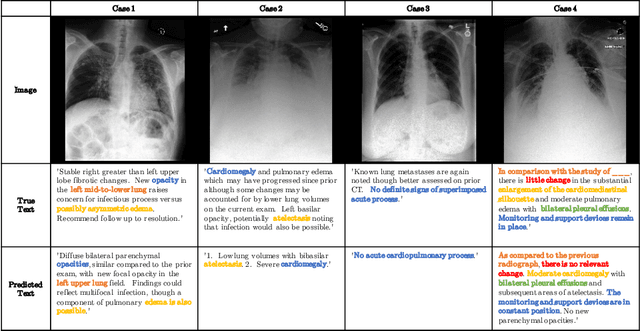
Abstract:Chest radiographs are one of the most common diagnostic modalities in clinical routine. It can be done cheaply, requires minimal equipment, and the image can be diagnosed by every radiologists. However, the number of chest radiographs obtained on a daily basis can easily overwhelm the available clinical capacities. We propose RATCHET: RAdiological Text Captioning for Human Examined Thoraces. RATCHET is a CNN-RNN-based medical transformer that is trained end-to-end. It is capable of extracting image features from chest radiographs, and generates medically accurate text reports that fit seamlessly into clinical work flows. The model is evaluated for its natural language generation ability using common metrics from NLP literature, as well as its medically accuracy through a surrogate report classification task. The model is available for download at: http://www.github.com/farrell236/RATCHET.
Deep Lesion Graphs in the Wild: Relationship Learning and Organization of Significant Radiology Image Findings in a Diverse Large-scale Lesion Database
Jul 28, 2018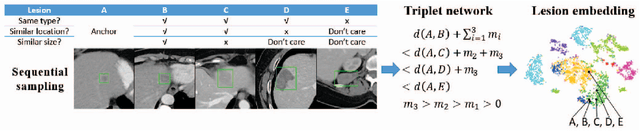
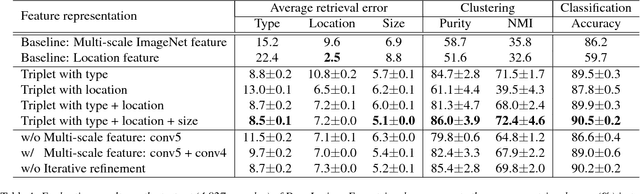
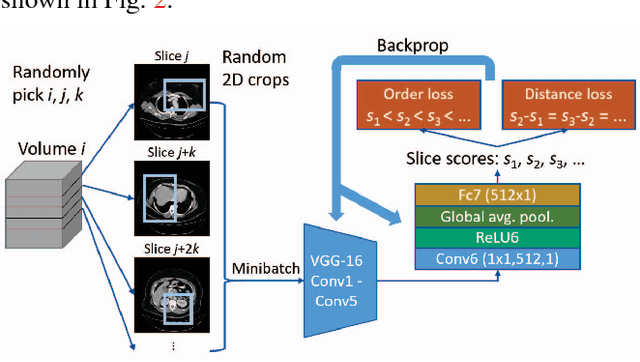
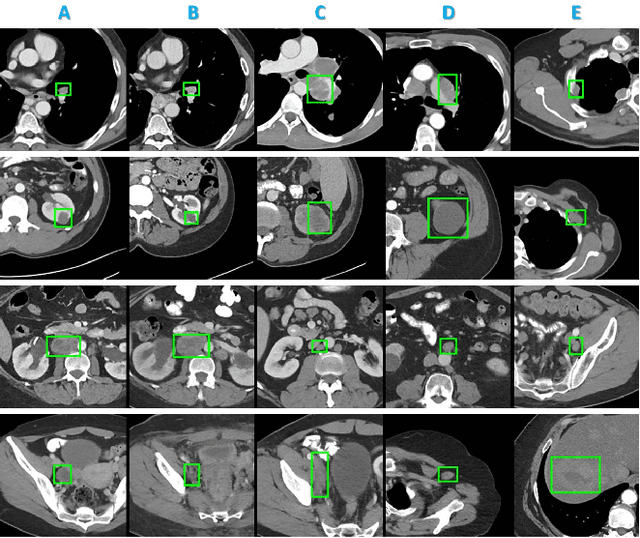
Abstract:Radiologists in their daily work routinely find and annotate significant abnormalities on a large number of radiology images. Such abnormalities, or lesions, have collected over years and stored in hospitals' picture archiving and communication systems. However, they are basically unsorted and lack semantic annotations like type and location. In this paper, we aim to organize and explore them by learning a deep feature representation for each lesion. A large-scale and comprehensive dataset, DeepLesion, is introduced for this task. DeepLesion contains bounding boxes and size measurements of over 32K lesions. To model their similarity relationship, we leverage multiple supervision information including types, self-supervised location coordinates and sizes. They require little manual annotation effort but describe useful attributes of the lesions. Then, a triplet network is utilized to learn lesion embeddings with a sequential sampling strategy to depict their hierarchical similarity structure. Experiments show promising qualitative and quantitative results on lesion retrieval, clustering, and classification. The learned embeddings can be further employed to build a lesion graph for various clinically useful applications. We propose algorithms for intra-patient lesion matching and missing annotation mining. Experimental results validate their effectiveness.
NegBio: a high-performance tool for negation and uncertainty detection in radiology reports
Dec 27, 2017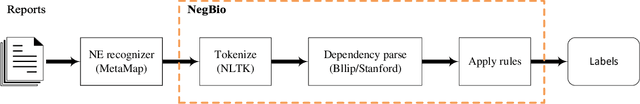
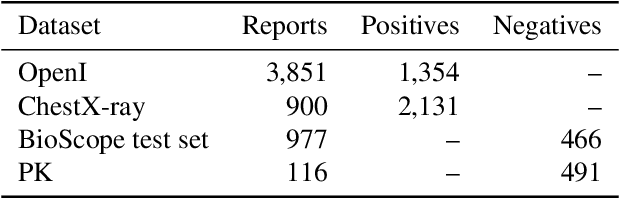
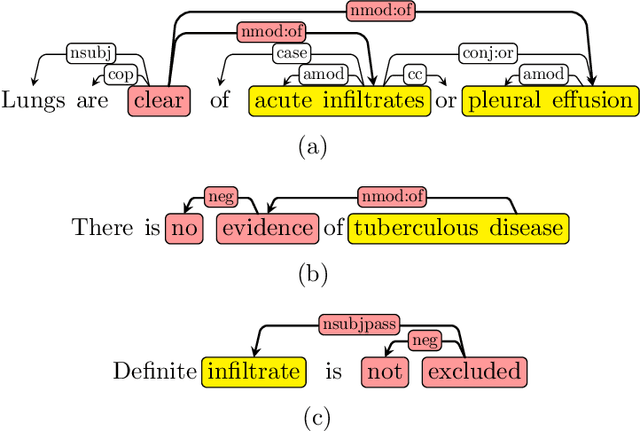
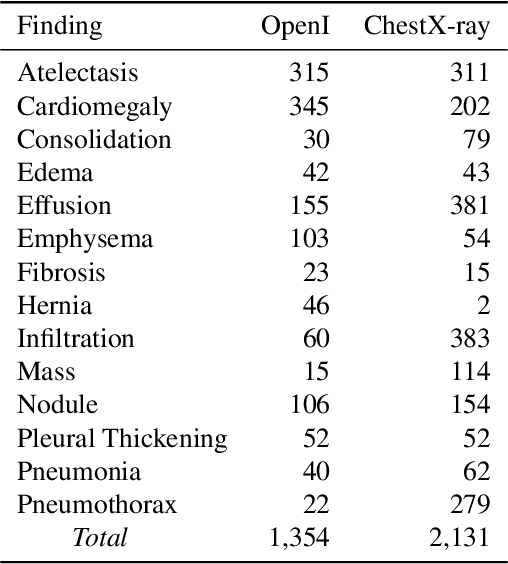
Abstract:Negative and uncertain medical findings are frequent in radiology reports, but discriminating them from positive findings remains challenging for information extraction. Here, we propose a new algorithm, NegBio, to detect negative and uncertain findings in radiology reports. Unlike previous rule-based methods, NegBio utilizes patterns on universal dependencies to identify the scope of triggers that are indicative of negation or uncertainty. We evaluated NegBio on four datasets, including two public benchmarking corpora of radiology reports, a new radiology corpus that we annotated for this work, and a public corpus of general clinical texts. Evaluation on these datasets demonstrates that NegBio is highly accurate for detecting negative and uncertain findings and compares favorably to a widely-used state-of-the-art system NegEx (an average of 9.5% improvement in precision and 5.1% in F1-score).
Unsupervised Category Discovery via Looped Deep Pseudo-Task Optimization Using a Large Scale Radiology Image Database
Mar 25, 2016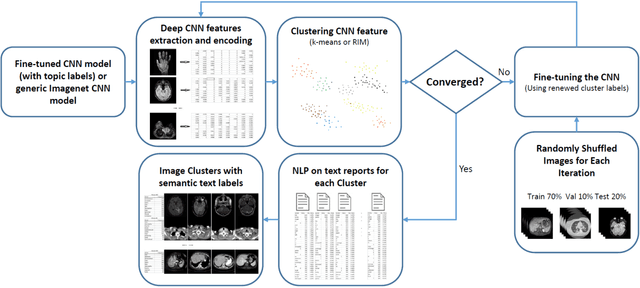
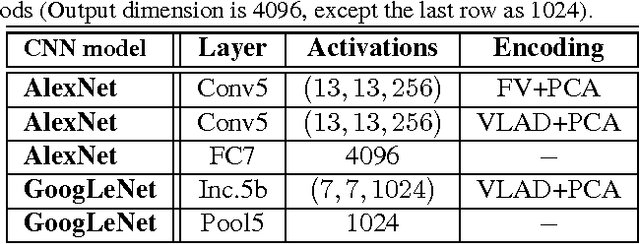
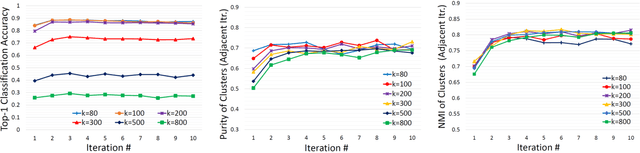
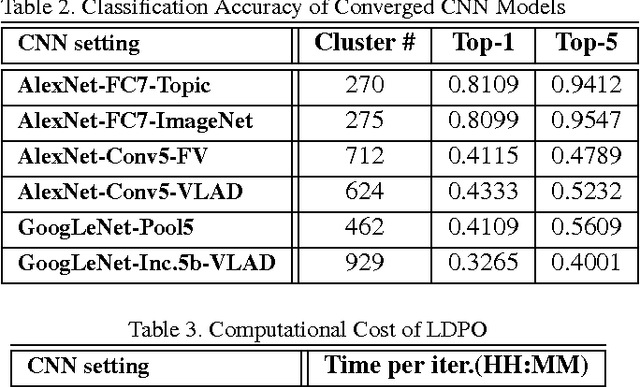
Abstract:Obtaining semantic labels on a large scale radiology image database (215,786 key images from 61,845 unique patients) is a prerequisite yet bottleneck to train highly effective deep convolutional neural network (CNN) models for image recognition. Nevertheless, conventional methods for collecting image labels (e.g., Google search followed by crowd-sourcing) are not applicable due to the formidable difficulties of medical annotation tasks for those who are not clinically trained. This type of image labeling task remains non-trivial even for radiologists due to uncertainty and possible drastic inter-observer variation or inconsistency. In this paper, we present a looped deep pseudo-task optimization procedure for automatic category discovery of visually coherent and clinically semantic (concept) clusters. Our system can be initialized by domain-specific (CNN trained on radiology images and text report derived labels) or generic (ImageNet based) CNN models. Afterwards, a sequence of pseudo-tasks are exploited by the looped deep image feature clustering (to refine image labels) and deep CNN training/classification using new labels (to obtain more task representative deep features). Our method is conceptually simple and based on the hypothesized "convergence" of better labels leading to better trained CNN models which in turn feed more effective deep image features to facilitate more meaningful clustering/labels. We have empirically validated the convergence and demonstrated promising quantitative and qualitative results. Category labels of significantly higher quality than those in previous work are discovered. This allows for further investigation of the hierarchical semantic nature of the given large-scale radiology image database.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge