Pritam Mukherjee
Text Embedded Swin-UMamba for DeepLesion Segmentation
Aug 08, 2025Abstract:Segmentation of lesions on CT enables automatic measurement for clinical assessment of chronic diseases (e.g., lymphoma). Integrating large language models (LLMs) into the lesion segmentation workflow offers the potential to combine imaging features with descriptions of lesion characteristics from the radiology reports. In this study, we investigate the feasibility of integrating text into the Swin-UMamba architecture for the task of lesion segmentation. The publicly available ULS23 DeepLesion dataset was used along with short-form descriptions of the findings from the reports. On the test dataset, a high Dice Score of 82% and low Hausdorff distance of 6.58 (pixels) was obtained for lesion segmentation. The proposed Text-Swin-UMamba model outperformed prior approaches: 37% improvement over the LLM-driven LanGuideMedSeg model (p < 0.001),and surpassed the purely image-based xLSTM-UNet and nnUNet models by 1.74% and 0.22%, respectively. The dataset and code can be accessed at https://github.com/ruida/LLM-Swin-UMamba
Benchmarking Multi-Organ Segmentation Tools for Multi-Parametric T1-weighted Abdominal MRI
Apr 10, 2025Abstract:The segmentation of multiple organs in multi-parametric MRI studies is critical for many applications in radiology, such as correlating imaging biomarkers with disease status (e.g., cirrhosis, diabetes). Recently, three publicly available tools, such as MRSegmentator (MRSeg), TotalSegmentator MRI (TS), and TotalVibeSegmentator (VIBE), have been proposed for multi-organ segmentation in MRI. However, the performance of these tools on specific MRI sequence types has not yet been quantified. In this work, a subset of 40 volumes from the public Duke Liver Dataset was curated. The curated dataset contained 10 volumes each from the pre-contrast fat saturated T1, arterial T1w, venous T1w, and delayed T1w phases, respectively. Ten abdominal structures were manually annotated in these volumes. Next, the performance of the three public tools was benchmarked on this curated dataset. The results indicated that MRSeg obtained a Dice score of 80.7 $\pm$ 18.6 and Hausdorff Distance (HD) error of 8.9 $\pm$ 10.4 mm. It fared the best ($p < .05$) across the different sequence types in contrast to TS and VIBE.
Leveraging Anatomical Priors for Automated Pancreas Segmentation on Abdominal CT
Apr 09, 2025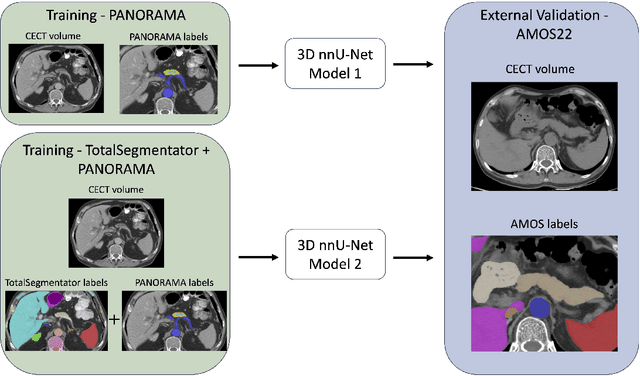


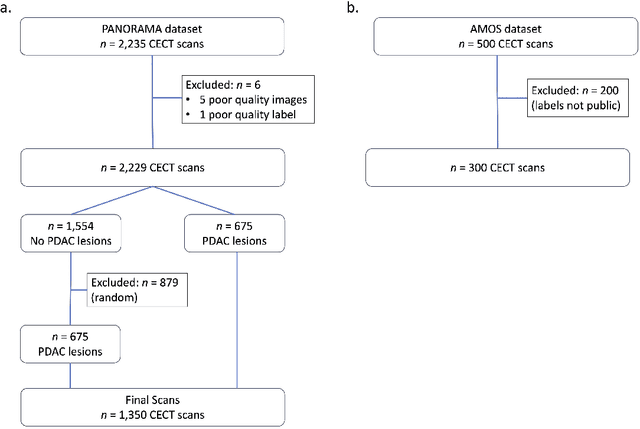
Abstract:An accurate segmentation of the pancreas on CT is crucial to identify pancreatic pathologies and extract imaging-based biomarkers. However, prior research on pancreas segmentation has primarily focused on modifying the segmentation model architecture or utilizing pre- and post-processing techniques. In this article, we investigate the utility of anatomical priors to enhance the segmentation performance of the pancreas. Two 3D full-resolution nnU-Net models were trained, one with 8 refined labels from the public PANORAMA dataset, and another that combined them with labels derived from the public TotalSegmentator (TS) tool. The addition of anatomical priors resulted in a 6\% increase in Dice score ($p < .001$) and a 36.5 mm decrease in Hausdorff distance for pancreas segmentation ($p < .001$). Moreover, the pancreas was always detected when anatomy priors were used, whereas there were 8 instances of failed detections without their use. The use of anatomy priors shows promise for pancreas segmentation and subsequent derivation of imaging biomarkers.
LEAVS: An LLM-based Labeler for Abdominal CT Supervision
Mar 17, 2025Abstract:Extracting structured labels from radiology reports has been employed to create vision models to simultaneously detect several types of abnormalities. However, existing works focus mainly on the chest region. Few works have been investigated on abdominal radiology reports due to more complex anatomy and a wider range of pathologies in the abdomen. We propose LEAVS (Large language model Extractor for Abdominal Vision Supervision). This labeler can annotate the certainty of presence and the urgency of seven types of abnormalities for nine abdominal organs on CT radiology reports. To ensure broad coverage, we chose abnormalities that encompass most of the finding types from CT reports. Our approach employs a specialized chain-of-thought prompting strategy for a locally-run LLM using sentence extraction and multiple-choice questions in a tree-based decision system. We demonstrate that the LLM can extract several abnormality types across abdominal organs with an average F1 score of 0.89, significantly outperforming competing labelers and humans. Additionally, we show that extraction of urgency labels achieved performance comparable to human annotations. Finally, we demonstrate that the abnormality labels contain valuable information for training a single vision model that classifies several organs as normal or abnormal. We release our code and structured annotations for a public CT dataset containing over 1,000 CT volumes.
Efficient 2D CT Foundation Model for Contrast Phase Classification
Jan 23, 2025Abstract:Purpose: The purpose of this study is to harness the efficiency of a 2D foundation model to develop a robust phase classifier that is resilient to domain shifts. Materials and Methods: This retrospective study utilized three public datasets from separate institutions. A 2D foundation model was trained on the DeepLesion dataset (mean age: 51.2, s.d.: 17.6; 2398 males) to generate embeddings from 2D CT slices for downstream contrast phase classification. The classifier was trained on the VinDr Multiphase dataset and externally validated on the WAW-TACE dataset. The 2D model was also compared to three 3D supervised models. Results: On the VinDr dataset (146 male, 63 female, 56 unidentified), the model achieved near-perfect AUROC scores and F1 scores of 99.2%, 94.2%, and 93.1% for non-contrast, arterial, and venous phases, respectively. The `Other' category scored lower (F1: 73.4%) due to combining multiple contrast phases into one class. On the WAW-TACE dataset (mean age: 66.1, s.d.: 10.0; 185 males), the model showed strong performance with AUROCs of 91.0% and 85.6%, and F1 scores of 87.3% and 74.1% for non-contrast and arterial phases. Venous phase performance was lower, with AUROC and F1 scores of 81.7% and 70.2% respectively, due to label mismatches. Compared to 3D supervised models, the approach trained faster, performed as well or better, and showed greater robustness to domain shifts. Conclusion: The robustness of the 2D Foundation model may be potentially useful for automation of hanging protocols and data orchestration for clinical deployment of AI algorithms.
MRISegmentator-Abdomen: A Fully Automated Multi-Organ and Structure Segmentation Tool for T1-weighted Abdominal MRI
May 09, 2024



Abstract:Background: Segmentation of organs and structures in abdominal MRI is useful for many clinical applications, such as disease diagnosis and radiotherapy. Current approaches have focused on delineating a limited set of abdominal structures (13 types). To date, there is no publicly available abdominal MRI dataset with voxel-level annotations of multiple organs and structures. Consequently, a segmentation tool for multi-structure segmentation is also unavailable. Methods: We curated a T1-weighted abdominal MRI dataset consisting of 195 patients who underwent imaging at National Institutes of Health (NIH) Clinical Center. The dataset comprises of axial pre-contrast T1, arterial, venous, and delayed phases for each patient, thereby amounting to a total of 780 series (69,248 2D slices). Each series contains voxel-level annotations of 62 abdominal organs and structures. A 3D nnUNet model, dubbed as MRISegmentator-Abdomen (MRISegmentator in short), was trained on this dataset, and evaluation was conducted on an internal test set and two large external datasets: AMOS22 and Duke Liver. The predicted segmentations were compared against the ground-truth using the Dice Similarity Coefficient (DSC) and Normalized Surface Distance (NSD). Findings: MRISegmentator achieved an average DSC of 0.861$\pm$0.170 and a NSD of 0.924$\pm$0.163 in the internal test set. On the AMOS22 dataset, MRISegmentator attained an average DSC of 0.829$\pm$0.133 and a NSD of 0.908$\pm$0.067. For the Duke Liver dataset, an average DSC of 0.933$\pm$0.015 and a NSD of 0.929$\pm$0.021 was obtained. Interpretation: The proposed MRISegmentator provides automatic, accurate, and robust segmentations of 62 organs and structures in T1-weighted abdominal MRI sequences. The tool has the potential to accelerate research on various clinical topics, such as abnormality detection, radiotherapy, disease classification among others.
Decomposing Vision-based LLM Predictions for Auto-Evaluation with GPT-4
Mar 08, 2024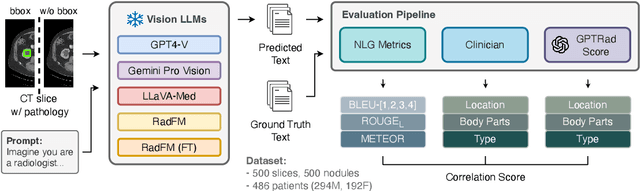

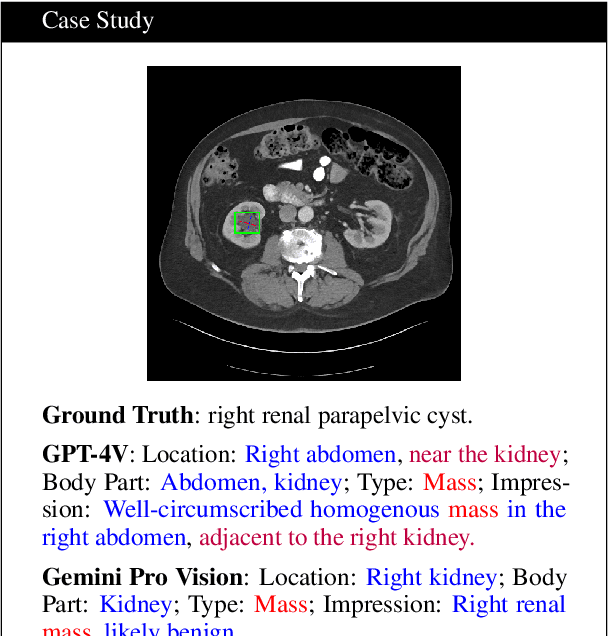

Abstract:The volume of CT exams being done in the world has been rising every year, which has led to radiologist burn-out. Large Language Models (LLMs) have the potential to reduce their burden, but their adoption in the clinic depends on radiologist trust, and easy evaluation of generated content. Presently, many automated methods are available to evaluate the reports generated for chest radiographs, but such an approach is not available for CT presently. In this paper, we propose a novel evaluation framework to judge the capabilities of vision-language LLMs in generating accurate summaries of CT-based abnormalities. CT slices containing an abnormality (e.g., lesion) were input to a vision-based LLM (GPT-4V, LLaVA-Med, and RadFM), and it generated a free-text summary of the predicted characteristics of the abnormality. Next, a GPT-4 model decomposed the summary into specific aspects (body part, location, type, and attributes), automatically evaluated the characteristics against the ground-truth, and generated a score for each aspect based on its clinical relevance and factual accuracy. These scores were then contrasted against those obtained from a clinician, and a high correlation ( 85%, p < .001) was observed. Although GPT-4V outperformed other models in our evaluation, it still requires overall improvement. Our evaluation method offers valuable insights into the specific areas that need the most enhancement, guiding future development in this field.
Enhancing chest X-ray datasets with privacy-preserving large language models and multi-type annotations: a data-driven approach for improved classification
Mar 06, 2024Abstract:In chest X-ray (CXR) image analysis, rule-based systems are usually employed to extract labels from reports, but concerns exist about label quality. These datasets typically offer only presence labels, sometimes with binary uncertainty indicators, which limits their usefulness. In this work, we present MAPLEZ (Medical report Annotations with Privacy-preserving Large language model using Expeditious Zero shot answers), a novel approach leveraging a locally executable Large Language Model (LLM) to extract and enhance findings labels on CXR reports. MAPLEZ extracts not only binary labels indicating the presence or absence of a finding but also the location, severity, and radiologists' uncertainty about the finding. Over eight abnormalities from five test sets, we show that our method can extract these annotations with an increase of 5 percentage points (pp) in F1 score for categorical presence annotations and more than 30 pp increase in F1 score for the location annotations over competing labelers. Additionally, using these improved annotations in classification supervision, we demonstrate substantial advancements in model quality, with an increase of 1.7 pp in AUROC over models trained with annotations from the state-of-the-art approach. We share code and annotations.
Automated Classification of Body MRI Sequence Type Using Convolutional Neural Networks
Feb 12, 2024Abstract:Multi-parametric MRI of the body is routinely acquired for the identification of abnormalities and diagnosis of diseases. However, a standard naming convention for the MRI protocols and associated sequences does not exist due to wide variations in imaging practice at institutions and myriad MRI scanners from various manufacturers being used for imaging. The intensity distributions of MRI sequences differ widely as a result, and there also exists information conflicts related to the sequence type in the DICOM headers. At present, clinician oversight is necessary to ensure that the correct sequence is being read and used for diagnosis. This poses a challenge when specific series need to be considered for building a cohort for a large clinical study or for developing AI algorithms. In order to reduce clinician oversight and ensure the validity of the DICOM headers, we propose an automated method to classify the 3D MRI sequence acquired at the levels of the chest, abdomen, and pelvis. In our pilot work, our 3D DenseNet-121 model achieved an F1 score of 99.5% at differentiating 5 common MRI sequences obtained by three Siemens scanners (Aera, Verio, Biograph mMR). To the best of our knowledge, we are the first to develop an automated method for the 3D classification of MRI sequences in the chest, abdomen, and pelvis, and our work has outperformed the previous state-of-the-art MRI series classifiers.
Weakly Supervised Detection of Pheochromocytomas and Paragangliomas in CT
Feb 12, 2024Abstract:Pheochromocytomas and Paragangliomas (PPGLs) are rare adrenal and extra-adrenal tumors which have the potential to metastasize. For the management of patients with PPGLs, CT is the preferred modality of choice for precise localization and estimation of their progression. However, due to the myriad variations in size, morphology, and appearance of the tumors in different anatomical regions, radiologists are posed with the challenge of accurate detection of PPGLs. Since clinicians also need to routinely measure their size and track their changes over time across patient visits, manual demarcation of PPGLs is quite a time-consuming and cumbersome process. To ameliorate the manual effort spent for this task, we propose an automated method to detect PPGLs in CT studies via a proxy segmentation task. As only weak annotations for PPGLs in the form of prospectively marked 2D bounding boxes on an axial slice were available, we extended these 2D boxes into weak 3D annotations and trained a 3D full-resolution nnUNet model to directly segment PPGLs. We evaluated our approach on a dataset consisting of chest-abdomen-pelvis CTs of 255 patients with confirmed PPGLs. We obtained a precision of 70% and sensitivity of 64.1% with our proposed approach when tested on 53 CT studies. Our findings highlight the promising nature of detecting PPGLs via segmentation, and furthers the state-of-the-art in this exciting yet challenging area of rare cancer management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge