Ricardo Bigolin Lanfredi
LEAVS: An LLM-based Labeler for Abdominal CT Supervision
Mar 17, 2025Abstract:Extracting structured labels from radiology reports has been employed to create vision models to simultaneously detect several types of abnormalities. However, existing works focus mainly on the chest region. Few works have been investigated on abdominal radiology reports due to more complex anatomy and a wider range of pathologies in the abdomen. We propose LEAVS (Large language model Extractor for Abdominal Vision Supervision). This labeler can annotate the certainty of presence and the urgency of seven types of abnormalities for nine abdominal organs on CT radiology reports. To ensure broad coverage, we chose abnormalities that encompass most of the finding types from CT reports. Our approach employs a specialized chain-of-thought prompting strategy for a locally-run LLM using sentence extraction and multiple-choice questions in a tree-based decision system. We demonstrate that the LLM can extract several abnormality types across abdominal organs with an average F1 score of 0.89, significantly outperforming competing labelers and humans. Additionally, we show that extraction of urgency labels achieved performance comparable to human annotations. Finally, we demonstrate that the abnormality labels contain valuable information for training a single vision model that classifies several organs as normal or abnormal. We release our code and structured annotations for a public CT dataset containing over 1,000 CT volumes.
Enhancing chest X-ray datasets with privacy-preserving large language models and multi-type annotations: a data-driven approach for improved classification
Mar 06, 2024Abstract:In chest X-ray (CXR) image analysis, rule-based systems are usually employed to extract labels from reports, but concerns exist about label quality. These datasets typically offer only presence labels, sometimes with binary uncertainty indicators, which limits their usefulness. In this work, we present MAPLEZ (Medical report Annotations with Privacy-preserving Large language model using Expeditious Zero shot answers), a novel approach leveraging a locally executable Large Language Model (LLM) to extract and enhance findings labels on CXR reports. MAPLEZ extracts not only binary labels indicating the presence or absence of a finding but also the location, severity, and radiologists' uncertainty about the finding. Over eight abnormalities from five test sets, we show that our method can extract these annotations with an increase of 5 percentage points (pp) in F1 score for categorical presence annotations and more than 30 pp increase in F1 score for the location annotations over competing labelers. Additionally, using these improved annotations in classification supervision, we demonstrate substantial advancements in model quality, with an increase of 1.7 pp in AUROC over models trained with annotations from the state-of-the-art approach. We share code and annotations.
Localization supervision of chest x-ray classifiers using label-specific eye-tracking annotation
Jul 20, 2022



Abstract:Convolutional neural networks (CNNs) have been successfully applied to chest x-ray (CXR) images. Moreover, annotated bounding boxes have been shown to improve the interpretability of a CNN in terms of localizing abnormalities. However, only a few relatively small CXR datasets containing bounding boxes are available, and collecting them is very costly. Opportunely, eye-tracking (ET) data can be collected in a non-intrusive way during the clinical workflow of a radiologist. We use ET data recorded from radiologists while dictating CXR reports to train CNNs. We extract snippets from the ET data by associating them with the dictation of keywords and use them to supervise the localization of abnormalities. We show that this method improves a model's interpretability without impacting its image-level classification.
Comparing radiologists' gaze and saliency maps generated by interpretability methods for chest x-rays
Dec 22, 2021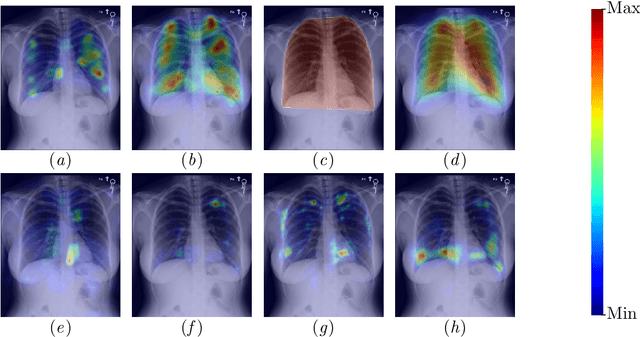
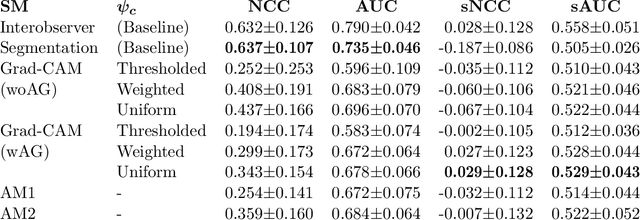
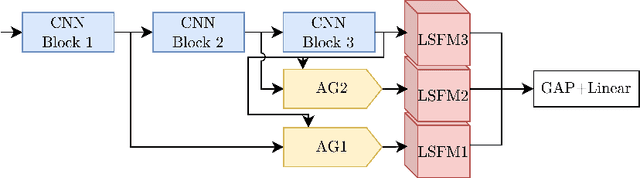
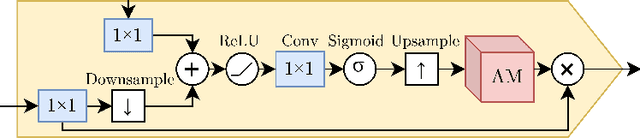
Abstract:The interpretability of medical image analysis models is considered a key research field. We use a dataset of eye-tracking data from five radiologists to compare the outputs of interpretability methods against the heatmaps representing where radiologists looked. We conduct a class-independent analysis of the saliency maps generated by two methods selected from the literature: Grad-CAM and attention maps from an attention-gated model. For the comparison, we use shuffled metrics, which avoid biases from fixation locations. We achieve scores comparable to an interobserver baseline in one shuffled metric, highlighting the potential of saliency maps from Grad-CAM to mimic a radiologist's attention over an image. We also divide the dataset into subsets to evaluate in which cases similarities are higher.
REFLACX, a dataset of reports and eye-tracking data for localization of abnormalities in chest x-rays
Sep 29, 2021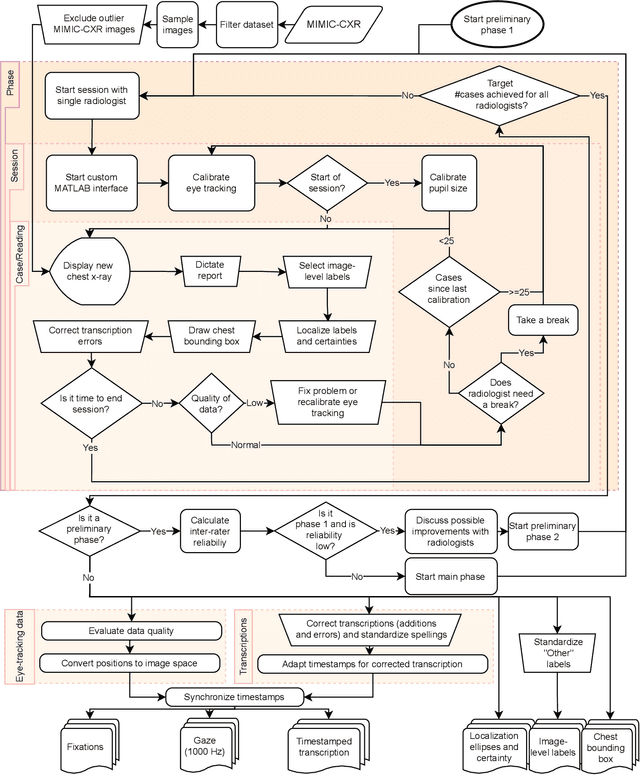



Abstract:Deep learning has shown recent success in classifying anomalies in chest x-rays, but datasets are still small compared to natural image datasets. Supervision of abnormality localization has been shown to improve trained models, partially compensating for dataset sizes. However, explicitly labeling these anomalies requires an expert and is very time-consuming. We propose a method for collecting implicit localization data using an eye tracker to capture gaze locations and a microphone to capture a dictation of a report, imitating the setup of a reading room, and potentially scalable for large datasets. The resulting REFLACX (Reports and Eye-Tracking Data for Localization of Abnormalities in Chest X-rays) dataset was labeled by five radiologists and contains 3,032 synchronized sets of eye-tracking data and timestamped report transcriptions. We also provide bounding boxes around lungs and heart and validation labels consisting of ellipses localizing abnormalities and image-level labels. Furthermore, a small subset of the data contains readings from all radiologists, allowing for the calculation of inter-rater scores.
Quantifying the Preferential Direction of the Model Gradient in Adversarial Training With Projected Gradient Descent
Sep 10, 2020



Abstract:Adversarial training, especially projected gradient descent (PGD), has been the most successful approach for improving robustness against adversarial attacks. After adversarial training, gradients of models with respect to their inputs are meaningful and interpretable by humans. However, the concept of interpretability is not mathematically well established, making it difficult to evaluate it quantitatively. We define interpretability as the alignment of the model gradient with the vector pointing toward the closest point of the support of the other class. We propose a method for measuring this alignment for binary classification problems, using generative adversarial model training to produce the smallest residual needed to change the class present in the image. We show that PGD-trained models are more interpretable than the baseline according to our definition, and our metric presents higher alignment values than a competing metric formulation. We also show that enforcing this alignment increases the robustness of models without adversarial training.
Interpretation of Disease Evidence for Medical Images Using Adversarial Deformation Fields
Jul 04, 2020



Abstract:The high complexity of deep learning models is associated with the difficulty of explaining what evidence they recognize as correlating with specific disease labels. This information is critical for building trust in models and finding their biases. Until now, automated deep learning visualization solutions have identified regions of images used by classifiers, but these solutions are too coarse, too noisy, or have a limited representation of the way images can change. We propose a novel method for formulating and presenting spatial explanations of disease evidence, called deformation field interpretation with generative adversarial networks (DeFI-GAN). An adversarially trained generator produces deformation fields that modify images of diseased patients to resemble images of healthy patients. We validate the method studying chronic obstructive pulmonary disease (COPD) evidence in chest x-rays (CXRs) and Alzheimer's disease (AD) evidence in brain MRIs. When extracting disease evidence in longitudinal data, we show compelling results against a baseline producing difference maps. DeFI-GAN also highlights disease biomarkers not found by previous methods and potential biases that may help in investigations of the dataset and of the adopted learning methods.
Adversarial regression training for visualizing the progression of chronic obstructive pulmonary disease with chest x-rays
Aug 27, 2019



Abstract:Knowledge of what spatial elements of medical images deep learning methods use as evidence is important for model interpretability, trustiness, and validation. There is a lack of such techniques for models in regression tasks. We propose a method, called visualization for regression with a generative adversarial network (VR-GAN), for formulating adversarial training specifically for datasets containing regression target values characterizing disease severity. We use a conditional generative adversarial network where the generator attempts to learn to shift the output of a regressor through creating disease effect maps that are added to the original images. Meanwhile, the regressor is trained to predict the original regression value for the modified images. A model trained with this technique learns to provide visualization for how the image would appear at different stages of the disease. We analyze our method in a dataset of chest x-rays associated with pulmonary function tests, used for diagnosing chronic obstructive pulmonary disease (COPD). For validation, we compute the difference of two registered x-rays of the same patient at different time points and correlate it to the generated disease effect map. The proposed method outperforms a technique based on classification and provides realistic-looking images, making modifications to images following what radiologists usually observe for this disease. Implementation code is available at https://github.com/ricbl/vrgan.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge