Lauren Kim
Unsupervised Joint Mining of Deep Features and Image Labels for Large-scale Radiology Image Categorization and Scene Recognition
Dec 27, 2017
Abstract:The recent rapid and tremendous success of deep convolutional neural networks (CNN) on many challenging computer vision tasks largely derives from the accessibility of the well-annotated ImageNet and PASCAL VOC datasets. Nevertheless, unsupervised image categorization (i.e., without the ground-truth labeling) is much less investigated, yet critically important and difficult when annotations are extremely hard to obtain in the conventional way of "Google Search" and crowd sourcing. We address this problem by presenting a looped deep pseudo-task optimization (LDPO) framework for joint mining of deep CNN features and image labels. Our method is conceptually simple and rests upon the hypothesized "convergence" of better labels leading to better trained CNN models which in turn feed more discriminative image representations to facilitate more meaningful clusters/labels. Our proposed method is validated in tackling two important applications: 1) Large-scale medical image annotation has always been a prohibitively expensive and easily-biased task even for well-trained radiologists. Significantly better image categorization results are achieved via our proposed approach compared to the previous state-of-the-art method. 2) Unsupervised scene recognition on representative and publicly available datasets with our proposed technique is examined. The LDPO achieves excellent quantitative scene classification results. On the MIT indoor scene dataset, it attains a clustering accuracy of 75.3%, compared to the state-of-the-art supervised classification accuracy of 81.0% (when both are based on the VGG-VD model).
Unsupervised Category Discovery via Looped Deep Pseudo-Task Optimization Using a Large Scale Radiology Image Database
Mar 25, 2016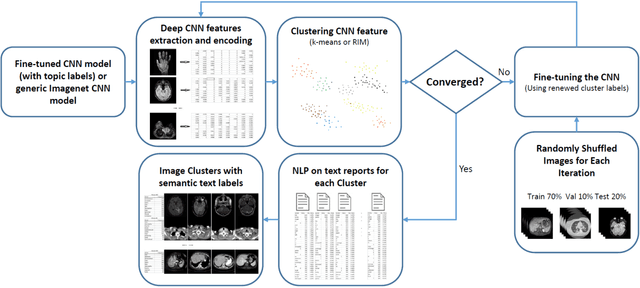
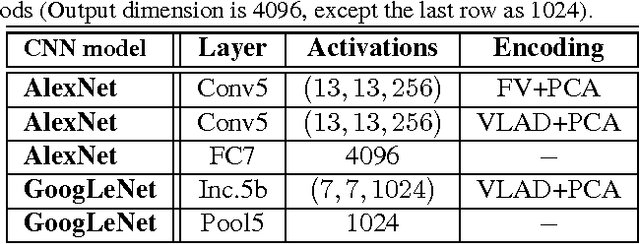
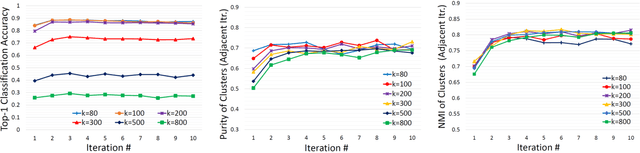
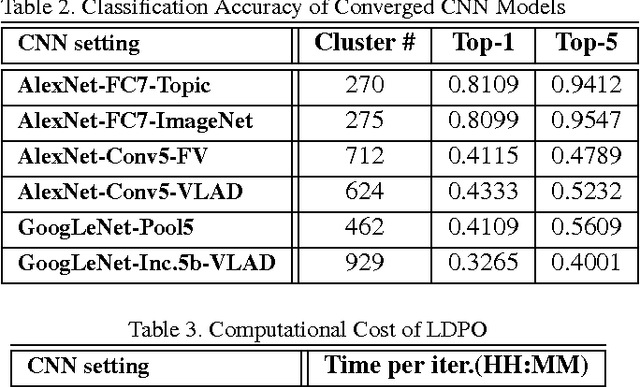
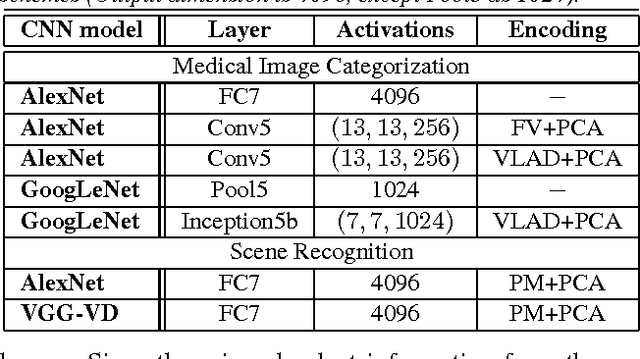
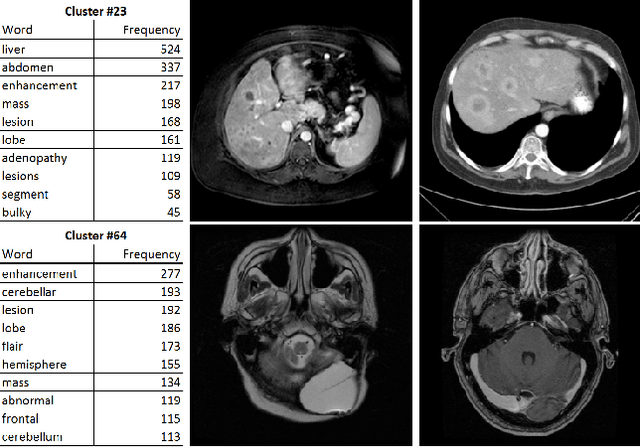
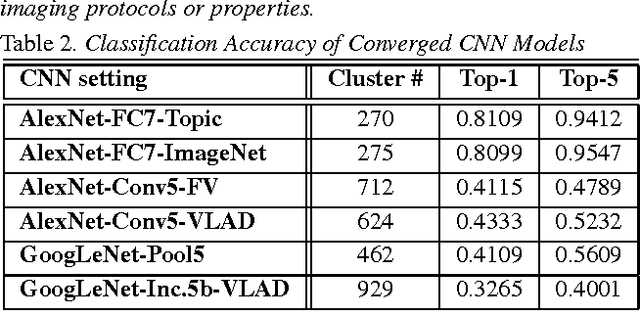
Abstract:Obtaining semantic labels on a large scale radiology image database (215,786 key images from 61,845 unique patients) is a prerequisite yet bottleneck to train highly effective deep convolutional neural network (CNN) models for image recognition. Nevertheless, conventional methods for collecting image labels (e.g., Google search followed by crowd-sourcing) are not applicable due to the formidable difficulties of medical annotation tasks for those who are not clinically trained. This type of image labeling task remains non-trivial even for radiologists due to uncertainty and possible drastic inter-observer variation or inconsistency. In this paper, we present a looped deep pseudo-task optimization procedure for automatic category discovery of visually coherent and clinically semantic (concept) clusters. Our system can be initialized by domain-specific (CNN trained on radiology images and text report derived labels) or generic (ImageNet based) CNN models. Afterwards, a sequence of pseudo-tasks are exploited by the looped deep image feature clustering (to refine image labels) and deep CNN training/classification using new labels (to obtain more task representative deep features). Our method is conceptually simple and based on the hypothesized "convergence" of better labels leading to better trained CNN models which in turn feed more effective deep image features to facilitate more meaningful clustering/labels. We have empirically validated the convergence and demonstrated promising quantitative and qualitative results. Category labels of significantly higher quality than those in previous work are discovered. This allows for further investigation of the hierarchical semantic nature of the given large-scale radiology image database.
Improving Computer-aided Detection using Convolutional Neural Networks and Random View Aggregation
Sep 15, 2015



Abstract:Automated computer-aided detection (CADe) in medical imaging has been an important tool in clinical practice and research. State-of-the-art methods often show high sensitivities but at the cost of high false-positives (FP) per patient rates. We design a two-tiered coarse-to-fine cascade framework that first operates a candidate generation system at sensitivities of $\sim$100% but at high FP levels. By leveraging existing CAD systems, coordinates of regions or volumes of interest (ROI or VOI) for lesion candidates are generated in this step and function as input for a second tier, which is our focus in this study. In this second stage, we generate $N$ 2D (two-dimensional) or 2.5D views via sampling through scale transformations, random translations and rotations with respect to each ROI's centroid coordinates. These random views are used to train deep convolutional neural network (ConvNet) classifiers. In testing, the trained ConvNets are employed to assign class (e.g., lesion, pathology) probabilities for a new set of $N$ random views that are then averaged at each ROI to compute a final per-candidate classification probability. This second tier behaves as a highly selective process to reject difficult false positives while preserving high sensitivities. The methods are evaluated on three different data sets with different numbers of patients: 59 patients for sclerotic metastases detection, 176 patients for lymph node detection, and 1,186 patients for colonic polyp detection. Experimental results show the ability of ConvNets to generalize well to different medical imaging CADe applications and scale elegantly to various data sets. Our proposed methods improve CADe performance markedly in all cases. CADe sensitivities improved from 57% to 70%, from 43% to 77% and from 58% to 75% at 3 FPs per patient for sclerotic metastases, lymph nodes and colonic polyps, respectively.
* 2D vs 2.5D vs 3D inputs and comparison to other standard classifiers such as SVM have been addressed by more experimentation and two completely new sections and figures. Results and Discussions have been updated accordingly
Interleaved Text/Image Deep Mining on a Large-Scale Radiology Database for Automated Image Interpretation
May 04, 2015



Abstract:Despite tremendous progress in computer vision, there has not been an attempt for machine learning on very large-scale medical image databases. We present an interleaved text/image deep learning system to extract and mine the semantic interactions of radiology images and reports from a national research hospital's Picture Archiving and Communication System. With natural language processing, we mine a collection of representative ~216K two-dimensional key images selected by clinicians for diagnostic reference, and match the images with their descriptions in an automated manner. Our system interleaves between unsupervised learning and supervised learning on document- and sentence-level text collections, to generate semantic labels and to predict them given an image. Given an image of a patient scan, semantic topics in radiology levels are predicted, and associated key-words are generated. Also, a number of frequent disease types are detected as present or absent, to provide more specific interpretation of a patient scan. This shows the potential of large-scale learning and prediction in electronic patient records available in most modern clinical institutions.
Anatomy-specific classification of medical images using deep convolutional nets
Apr 15, 2015



Abstract:Automated classification of human anatomy is an important prerequisite for many computer-aided diagnosis systems. The spatial complexity and variability of anatomy throughout the human body makes classification difficult. "Deep learning" methods such as convolutional networks (ConvNets) outperform other state-of-the-art methods in image classification tasks. In this work, we present a method for organ- or body-part-specific anatomical classification of medical images acquired using computed tomography (CT) with ConvNets. We train a ConvNet, using 4,298 separate axial 2D key-images to learn 5 anatomical classes. Key-images were mined from a hospital PACS archive, using a set of 1,675 patients. We show that a data augmentation approach can help to enrich the data set and improve classification performance. Using ConvNets and data augmentation, we achieve anatomy-specific classification error of 5.9 % and area-under-the-curve (AUC) values of an average of 0.998 in testing. We demonstrate that deep learning can be used to train very reliable and accurate classifiers that could initialize further computer-aided diagnosis.
* Presented at: 2015 IEEE International Symposium on Biomedical Imaging, April 16-19, 2015, New York Marriott at Brooklyn Bridge, NY, USA
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge