Huxin Gao
GeoLanG: Geometry-Aware Language-Guided Grasping with Unified RGB-D Multimodal Learning
Feb 04, 2026Abstract:Language-guided grasping has emerged as a promising paradigm for enabling robots to identify and manipulate target objects through natural language instructions, yet it remains highly challenging in cluttered or occluded scenes. Existing methods often rely on multi-stage pipelines that separate object perception and grasping, which leads to limited cross-modal fusion, redundant computation, and poor generalization in cluttered, occluded, or low-texture scenes. To address these limitations, we propose GeoLanG, an end-to-end multi-task framework built upon the CLIP architecture that unifies visual and linguistic inputs into a shared representation space for robust semantic alignment and improved generalization. To enhance target discrimination under occlusion and low-texture conditions, we explore a more effective use of depth information through the Depth-guided Geometric Module (DGGM), which converts depth into explicit geometric priors and injects them into the attention mechanism without additional computational overhead. In addition, we propose Adaptive Dense Channel Integration, which adaptively balances the contributions of multi-layer features to produce more discriminative and generalizable visual representations. Extensive experiments on the OCID-VLG dataset, as well as in both simulation and real-world hardware, demonstrate that GeoLanG enables precise and robust language-guided grasping in complex, cluttered environments, paving the way toward more reliable multimodal robotic manipulation in real-world human-centric settings.
EndoARSS: Adapting Spatially-Aware Foundation Model for Efficient Activity Recognition and Semantic Segmentation in Endoscopic Surgery
Jun 07, 2025Abstract:Endoscopic surgery is the gold standard for robotic-assisted minimally invasive surgery, offering significant advantages in early disease detection and precise interventions. However, the complexity of surgical scenes, characterized by high variability in different surgical activity scenarios and confused image features between targets and the background, presents challenges for surgical environment understanding. Traditional deep learning models often struggle with cross-activity interference, leading to suboptimal performance in each downstream task. To address this limitation, we explore multi-task learning, which utilizes the interrelated features between tasks to enhance overall task performance. In this paper, we propose EndoARSS, a novel multi-task learning framework specifically designed for endoscopy surgery activity recognition and semantic segmentation. Built upon the DINOv2 foundation model, our approach integrates Low-Rank Adaptation to facilitate efficient fine-tuning while incorporating Task Efficient Shared Low-Rank Adapters to mitigate gradient conflicts across diverse tasks. Additionally, we introduce the Spatially-Aware Multi-Scale Attention that enhances feature representation discrimination by enabling cross-spatial learning of global information. In order to evaluate the effectiveness of our framework, we present three novel datasets, MTLESD, MTLEndovis and MTLEndovis-Gen, tailored for endoscopic surgery scenarios with detailed annotations for both activity recognition and semantic segmentation tasks. Extensive experiments demonstrate that EndoARSS achieves remarkable performance across multiple benchmarks, significantly improving both accuracy and robustness in comparison to existing models. These results underscore the potential of EndoARSS to advance AI-driven endoscopic surgical systems, offering valuable insights for enhancing surgical safety and efficiency.
EndoVLA: Dual-Phase Vision-Language-Action Model for Autonomous Tracking in Endoscopy
May 21, 2025Abstract:In endoscopic procedures, autonomous tracking of abnormal regions and following circumferential cutting markers can significantly reduce the cognitive burden on endoscopists. However, conventional model-based pipelines are fragile for each component (e.g., detection, motion planning) requires manual tuning and struggles to incorporate high-level endoscopic intent, leading to poor generalization across diverse scenes. Vision-Language-Action (VLA) models, which integrate visual perception, language grounding, and motion planning within an end-to-end framework, offer a promising alternative by semantically adapting to surgeon prompts without manual recalibration. Despite their potential, applying VLA models to robotic endoscopy presents unique challenges due to the complex and dynamic anatomical environments of the gastrointestinal (GI) tract. To address this, we introduce EndoVLA, designed specifically for continuum robots in GI interventions. Given endoscopic images and surgeon-issued tracking prompts, EndoVLA performs three core tasks: (1) polyp tracking, (2) delineation and following of abnormal mucosal regions, and (3) adherence to circular markers during circumferential cutting. To tackle data scarcity and domain shifts, we propose a dual-phase strategy comprising supervised fine-tuning on our EndoVLA-Motion dataset and reinforcement fine-tuning with task-aware rewards. Our approach significantly improves tracking performance in endoscopy and enables zero-shot generalization in diverse scenes and complex sequential tasks.
ETSM: Automating Dissection Trajectory Suggestion and Confidence Map-Based Safety Margin Prediction for Robot-assisted Endoscopic Submucosal Dissection
Nov 28, 2024Abstract:Robot-assisted Endoscopic Submucosal Dissection (ESD) improves the surgical procedure by providing a more comprehensive view through advanced robotic instruments and bimanual operation, thereby enhancing dissection efficiency and accuracy. Accurate prediction of dissection trajectories is crucial for better decision-making, reducing intraoperative errors, and improving surgical training. Nevertheless, predicting these trajectories is challenging due to variable tumor margins and dynamic visual conditions. To address this issue, we create the ESD Trajectory and Confidence Map-based Safety Margin (ETSM) dataset with $1849$ short clips, focusing on submucosal dissection with a dual-arm robotic system. We also introduce a framework that combines optimal dissection trajectory prediction with a confidence map-based safety margin, providing a more secure and intelligent decision-making tool to minimize surgical risks for ESD procedures. Additionally, we propose the Regression-based Confidence Map Prediction Network (RCMNet), which utilizes a regression approach to predict confidence maps for dissection areas, thereby delineating various levels of safety margins. We evaluate our RCMNet using three distinct experimental setups: in-domain evaluation, robustness assessment, and out-of-domain evaluation. Experimental results show that our approach excels in the confidence map-based safety margin prediction task, achieving a mean absolute error (MAE) of only $3.18$. To the best of our knowledge, this is the first study to apply a regression approach for visual guidance concerning delineating varying safety levels of dissection areas. Our approach bridges gaps in current research by improving prediction accuracy and enhancing the safety of the dissection process, showing great clinical significance in practice.
PDZSeg: Adapting the Foundation Model for Dissection Zone Segmentation with Visual Prompts in Robot-assisted Endoscopic Submucosal Dissection
Nov 27, 2024Abstract:Purpose: Endoscopic surgical environments present challenges for dissection zone segmentation due to unclear boundaries between tissue types, leading to segmentation errors where models misidentify or overlook edges. This study aims to provide precise dissection zone suggestions during endoscopic submucosal dissection (ESD) procedures, enhancing ESD safety. Methods: We propose the Prompted-based Dissection Zone Segmentation (PDZSeg) model, designed to leverage diverse visual prompts such as scribbles and bounding boxes. By overlaying these prompts onto images and fine-tuning a foundational model on a specialized dataset, our approach improves segmentation performance and user experience through flexible input methods. Results: The PDZSeg model was validated using three experimental setups: in-domain evaluation, variability in visual prompt availability, and robustness assessment. Using the ESD-DZSeg dataset, results show that our method outperforms state-of-the-art segmentation approaches. This is the first study to integrate visual prompt design into dissection zone segmentation. Conclusion: The PDZSeg model effectively utilizes visual prompts to enhance segmentation performance and user experience, supported by the novel ESD-DZSeg dataset as a benchmark for dissection zone segmentation in ESD. Our work establishes a foundation for future research.
CoPESD: A Multi-Level Surgical Motion Dataset for Training Large Vision-Language Models to Co-Pilot Endoscopic Submucosal Dissection
Oct 10, 2024



Abstract:submucosal dissection (ESD) enables rapid resection of large lesions, minimizing recurrence rates and improving long-term overall survival. Despite these advantages, ESD is technically challenging and carries high risks of complications, necessitating skilled surgeons and precise instruments. Recent advancements in Large Visual-Language Models (LVLMs) offer promising decision support and predictive planning capabilities for robotic systems, which can augment the accuracy of ESD and reduce procedural risks. However, existing datasets for multi-level fine-grained ESD surgical motion understanding are scarce and lack detailed annotations. In this paper, we design a hierarchical decomposition of ESD motion granularity and introduce a multi-level surgical motion dataset (CoPESD) for training LVLMs as the robotic \textbf{Co}-\textbf{P}ilot of \textbf{E}ndoscopic \textbf{S}ubmucosal \textbf{D}issection. CoPESD includes 17,679 images with 32,699 bounding boxes and 88,395 multi-level motions, from over 35 hours of ESD videos for both robot-assisted and conventional surgeries. CoPESD enables granular analysis of ESD motions, focusing on the complex task of submucosal dissection. Extensive experiments on the LVLMs demonstrate the effectiveness of CoPESD in training LVLMs to predict following surgical robotic motions. As the first multimodal ESD motion dataset, CoPESD supports advanced research in ESD instruction-following and surgical automation. The dataset is available at \href{https://github.com/gkw0010/CoPESD}{https://github.com/gkw0010/CoPESD.}}
LighTDiff: Surgical Endoscopic Image Low-Light Enhancement with T-Diffusion
May 17, 2024



Abstract:Advances in endoscopy use in surgeries face challenges like inadequate lighting. Deep learning, notably the Denoising Diffusion Probabilistic Model (DDPM), holds promise for low-light image enhancement in the medical field. However, DDPMs are computationally demanding and slow, limiting their practical medical applications. To bridge this gap, we propose a lightweight DDPM, dubbed LighTDiff. It adopts a T-shape model architecture to capture global structural information using low-resolution images and gradually recover the details in subsequent denoising steps. We further prone the model to significantly reduce the model size while retaining performance. While discarding certain downsampling operations to save parameters leads to instability and low efficiency in convergence during the training, we introduce a Temporal Light Unit (TLU), a plug-and-play module, for more stable training and better performance. TLU associates time steps with denoised image features, establishing temporal dependencies of the denoising steps and improving denoising outcomes. Moreover, while recovering images using the diffusion model, potential spectral shifts were noted. We further introduce a Chroma Balancer (CB) to mitigate this issue. Our LighTDiff outperforms many competitive LLIE methods with exceptional computational efficiency.
Adapting SAM for Surgical Instrument Tracking and Segmentation in Endoscopic Submucosal Dissection Videos
Apr 16, 2024



Abstract:The precise tracking and segmentation of surgical instruments have led to a remarkable enhancement in the efficiency of surgical procedures. However, the challenge lies in achieving accurate segmentation of surgical instruments while minimizing the need for manual annotation and reducing the time required for the segmentation process. To tackle this, we propose a novel framework for surgical instrument segmentation and tracking. Specifically, with a tiny subset of frames for segmentation, we ensure accurate segmentation across the entire surgical video. Our method adopts a two-stage approach to efficiently segment videos. Initially, we utilize the Segment-Anything (SAM) model, which has been fine-tuned using the Low-Rank Adaptation (LoRA) on the EndoVis17 Dataset. The fine-tuned SAM model is applied to segment the initial frames of the video accurately. Subsequently, we deploy the XMem++ tracking algorithm to follow the annotated frames, thereby facilitating the segmentation of the entire video sequence. This workflow enables us to precisely segment and track objects within the video. Through extensive evaluation of the in-distribution dataset (EndoVis17) and the out-of-distribution datasets (EndoVis18 \& the endoscopic submucosal dissection surgery (ESD) dataset), our framework demonstrates exceptional accuracy and robustness, thus showcasing its potential to advance the automated robotic-assisted surgery.
OSSAR: Towards Open-Set Surgical Activity Recognition in Robot-assisted Surgery
Feb 10, 2024



Abstract:In the realm of automated robotic surgery and computer-assisted interventions, understanding robotic surgical activities stands paramount. Existing algorithms dedicated to surgical activity recognition predominantly cater to pre-defined closed-set paradigms, ignoring the challenges of real-world open-set scenarios. Such algorithms often falter in the presence of test samples originating from classes unseen during training phases. To tackle this problem, we introduce an innovative Open-Set Surgical Activity Recognition (OSSAR) framework. Our solution leverages the hyperspherical reciprocal point strategy to enhance the distinction between known and unknown classes in the feature space. Additionally, we address the issue of over-confidence in the closed set by refining model calibration, avoiding misclassification of unknown classes as known ones. To support our assertions, we establish an open-set surgical activity benchmark utilizing the public JIGSAWS dataset. Besides, we also collect a novel dataset on endoscopic submucosal dissection for surgical activity tasks. Extensive comparisons and ablation experiments on these datasets demonstrate the significant outperformance of our method over existing state-of-the-art approaches. Our proposed solution can effectively address the challenges of real-world surgical scenarios. Our code is publicly accessible at https://github.com/longbai1006/OSSAR.
A Miniature 3-DoF Flexible Parallel Robotic Wrist Using NiTi Wires for Gastrointestinal Endoscopic Surgery
Jul 11, 2022
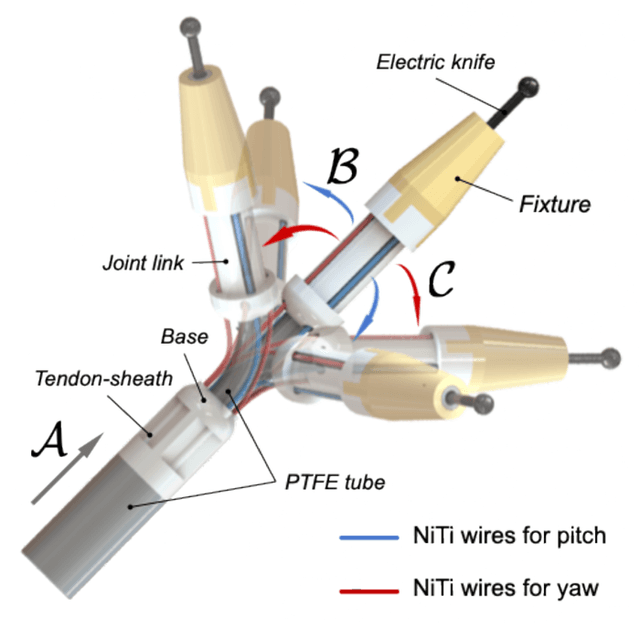
Abstract:Gastrointestinal endoscopic surgery (GES) has high requirements for instruments' size and distal dexterity, because of the narrow endoscopic channel and long, tortuous human gastrointestinal tract. This paper utilized Nickel-Titanium (NiTi) wires to develop a miniature 3-DoF (pitch-yaw-translation) flexible parallel robotic wrist (FPRW). Additionally, we assembled an electric knife on the wrist's connection interface and then teleoperated it to perform an endoscopic submucosal dissection (ESD) on porcine stomachs. The effective performance in each ESD workflow proves that the designed FPRW has sufficient workspace, high distal dexterity, and high positioning accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge