Erjian Guo
DiN: Diffusion Model for Robust Medical VQA with Semantic Noisy Labels
Mar 24, 2025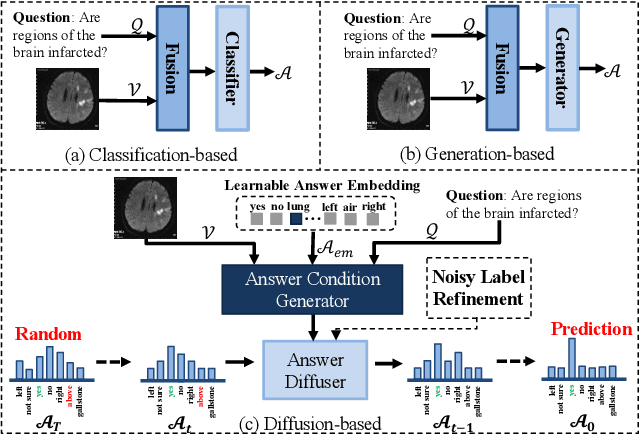
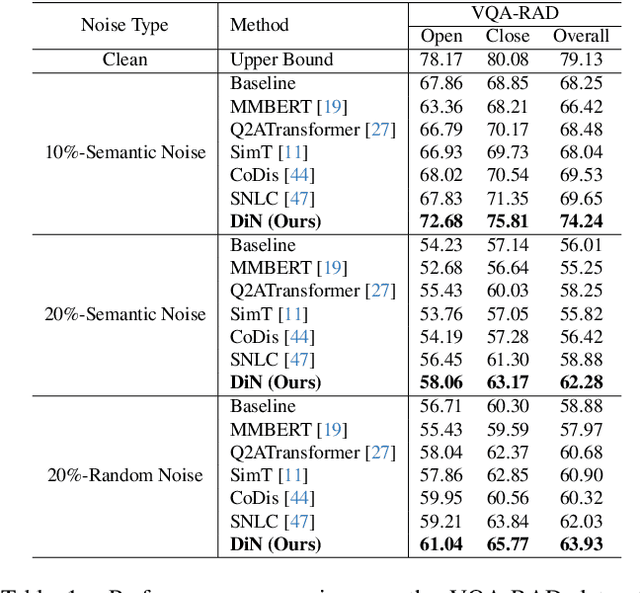
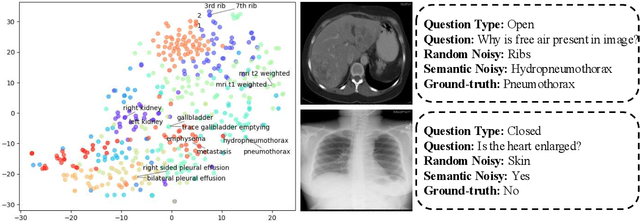
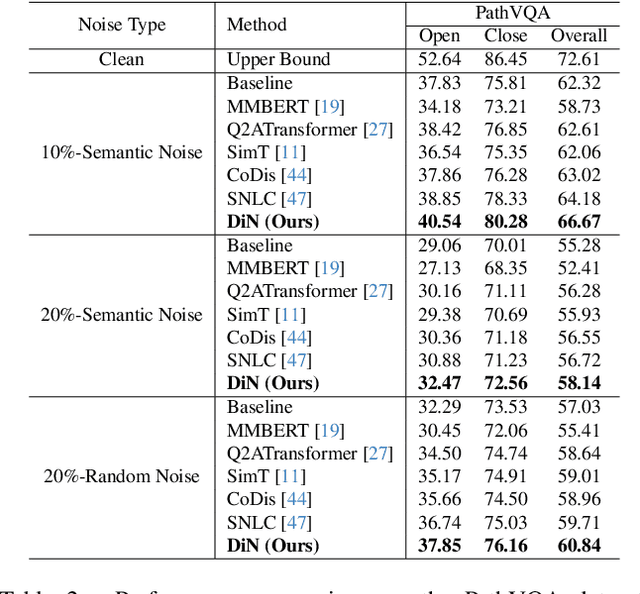
Abstract:Medical Visual Question Answering (Med-VQA) systems benefit the interpretation of medical images containing critical clinical information. However, the challenge of noisy labels and limited high-quality datasets remains underexplored. To address this, we establish the first benchmark for noisy labels in Med-VQA by simulating human mislabeling with semantically designed noise types. More importantly, we introduce the DiN framework, which leverages a diffusion model to handle noisy labels in Med-VQA. Unlike the dominant classification-based VQA approaches that directly predict answers, our Answer Diffuser (AD) module employs a coarse-to-fine process, refining answer candidates with a diffusion model for improved accuracy. The Answer Condition Generator (ACG) further enhances this process by generating task-specific conditional information via integrating answer embeddings with fused image-question features. To address label noise, our Noisy Label Refinement(NLR) module introduces a robust loss function and dynamic answer adjustment to further boost the performance of the AD module.
Imbalanced Medical Image Segmentation with Pixel-dependent Noisy Labels
Jan 12, 2025



Abstract:Accurate medical image segmentation is often hindered by noisy labels in training data, due to the challenges of annotating medical images. Prior research works addressing noisy labels tend to make class-dependent assumptions, overlooking the pixel-dependent nature of most noisy labels. Furthermore, existing methods typically apply fixed thresholds to filter out noisy labels, risking the removal of minority classes and consequently degrading segmentation performance. To bridge these gaps, our proposed framework, Collaborative Learning with Curriculum Selection (CLCS), addresses pixel-dependent noisy labels with class imbalance. CLCS advances the existing works by i) treating noisy labels as pixel-dependent and addressing them through a collaborative learning framework, and ii) employing a curriculum dynamic thresholding approach adapting to model learning progress to select clean data samples to mitigate the class imbalance issue, and iii) applying a noise balance loss to noisy data samples to improve data utilization instead of discarding them outright. Specifically, our CLCS contains two modules: Curriculum Noisy Label Sample Selection (CNS) and Noise Balance Loss (NBL). In the CNS module, we designed a two-branch network with discrepancy loss for collaborative learning so that different feature representations of the same instance could be extracted from distinct views and used to vote the class probabilities of pixels. Besides, a curriculum dynamic threshold is adopted to select clean-label samples through probability voting. In the NBL module, instead of directly dropping the suspiciously noisy labels, we further adopt a robust loss to leverage such instances to boost the performance.
LighTDiff: Surgical Endoscopic Image Low-Light Enhancement with T-Diffusion
May 17, 2024



Abstract:Advances in endoscopy use in surgeries face challenges like inadequate lighting. Deep learning, notably the Denoising Diffusion Probabilistic Model (DDPM), holds promise for low-light image enhancement in the medical field. However, DDPMs are computationally demanding and slow, limiting their practical medical applications. To bridge this gap, we propose a lightweight DDPM, dubbed LighTDiff. It adopts a T-shape model architecture to capture global structural information using low-resolution images and gradually recover the details in subsequent denoising steps. We further prone the model to significantly reduce the model size while retaining performance. While discarding certain downsampling operations to save parameters leads to instability and low efficiency in convergence during the training, we introduce a Temporal Light Unit (TLU), a plug-and-play module, for more stable training and better performance. TLU associates time steps with denoised image features, establishing temporal dependencies of the denoising steps and improving denoising outcomes. Moreover, while recovering images using the diffusion model, potential spectral shifts were noted. We further introduce a Chroma Balancer (CB) to mitigate this issue. Our LighTDiff outperforms many competitive LLIE methods with exceptional computational efficiency.
Clean Label Disentangling for Medical Image Segmentation with Noisy Labels
Nov 28, 2023Abstract:Current methods focusing on medical image segmentation suffer from incorrect annotations, which is known as the noisy label issue. Most medical image segmentation with noisy labels methods utilize either noise transition matrix, noise-robust loss functions or pseudo-labeling methods, while none of the current research focuses on clean label disentanglement. We argue that the main reason is that the severe class-imbalanced issue will lead to the inaccuracy of the selected ``clean'' labels, thus influencing the robustness of the model against the noises. In this work, we come up with a simple but efficient class-balanced sampling strategy to tackle the class-imbalanced problem, which enables our newly proposed clean label disentangling framework to successfully select clean labels from the given label sets and encourages the model to learn from the correct annotations. However, such a method will filter out too many annotations which may also contain useful information. Therefore, we further extend our clean label disentangling framework to a new noisy feature-aided clean label disentangling framework, which takes the full annotations into utilization to learn more semantics. Extensive experiments have validated the effectiveness of our methods, where our methods achieve new state-of-the-art performance. Our code is available at https://github.com/xiaoyao3302/2BDenoise.
Bridging Synthetic and Real Images: a Transferable and Multiple Consistency aided Fundus Image Enhancement Framework
Feb 23, 2023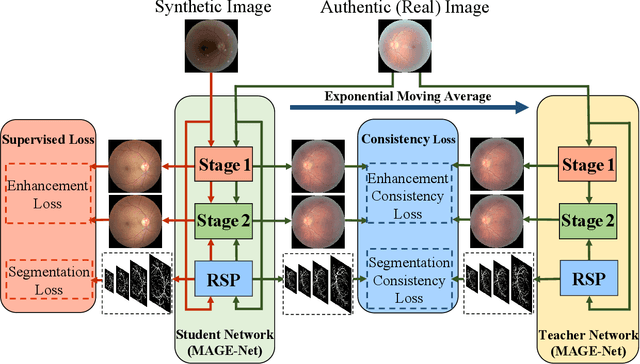
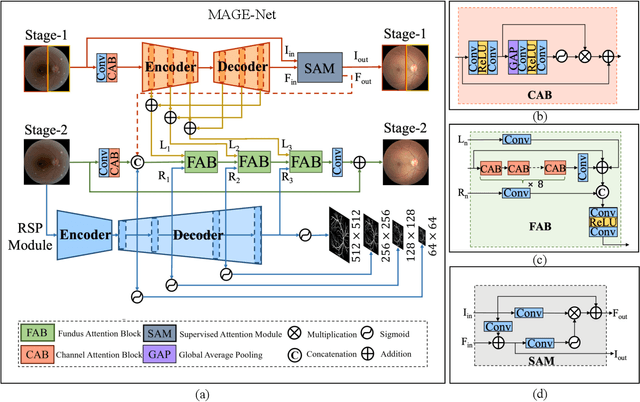
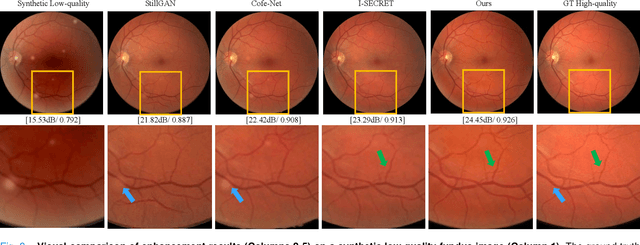
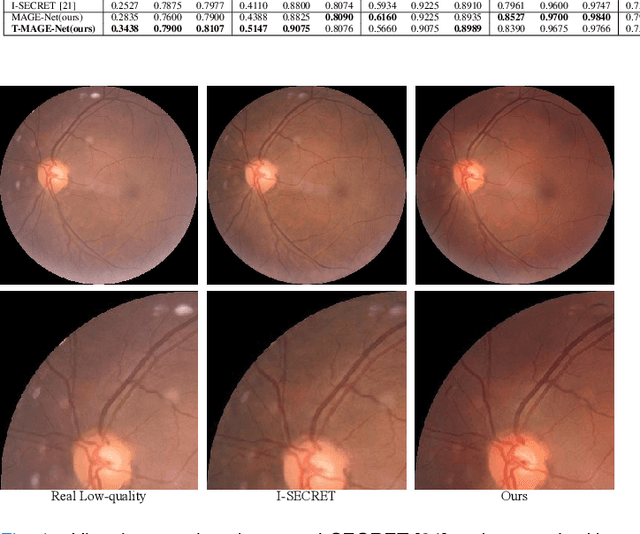
Abstract:Deep learning based image enhancement models have largely improved the readability of fundus images in order to decrease the uncertainty of clinical observations and the risk of misdiagnosis. However, due to the difficulty of acquiring paired real fundus images at different qualities, most existing methods have to adopt synthetic image pairs as training data. The domain shift between the synthetic and the real images inevitably hinders the generalization of such models on clinical data. In this work, we propose an end-to-end optimized teacher-student framework to simultaneously conduct image enhancement and domain adaptation. The student network uses synthetic pairs for supervised enhancement, and regularizes the enhancement model to reduce domain-shift by enforcing teacher-student prediction consistency on the real fundus images without relying on enhanced ground-truth. Moreover, we also propose a novel multi-stage multi-attention guided enhancement network (MAGE-Net) as the backbones of our teacher and student network. Our MAGE-Net utilizes multi-stage enhancement module and retinal structure preservation module to progressively integrate the multi-scale features and simultaneously preserve the retinal structures for better fundus image quality enhancement. Comprehensive experiments on both real and synthetic datasets demonstrate that our framework outperforms the baseline approaches. Moreover, our method also benefits the downstream clinical tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge