Hassan M. Fathallah-Shaykh
The Brain Tumor Segmentation Challenge 2023: Brain MR Image Synthesis for Tumor Segmentation
May 20, 2023
Abstract:Automated brain tumor segmentation methods are well established, reaching performance levels with clear clinical utility. Most algorithms require four input magnetic resonance imaging (MRI) modalities, typically T1-weighted images with and without contrast enhancement, T2-weighted images, and FLAIR images. However, some of these sequences are often missing in clinical practice, e.g., because of time constraints and/or image artifacts (such as patient motion). Therefore, substituting missing modalities to recover segmentation performance in these scenarios is highly desirable and necessary for the more widespread adoption of such algorithms in clinical routine. In this work, we report the set-up of the Brain MR Image Synthesis Benchmark (BraSyn), organized in conjunction with the Medical Image Computing and Computer-Assisted Intervention (MICCAI) 2023. The objective of the challenge is to benchmark image synthesis methods that realistically synthesize missing MRI modalities given multiple available images to facilitate automated brain tumor segmentation pipelines. The image dataset is multi-modal and diverse, created in collaboration with various hospitals and research institutions.
EvalAttAI: A Holistic Approach to Evaluating Attribution Maps in Robust and Non-Robust Models
Mar 15, 2023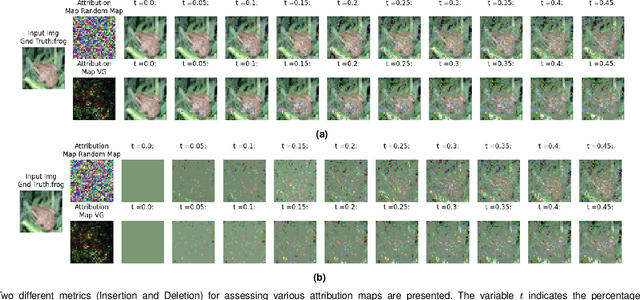
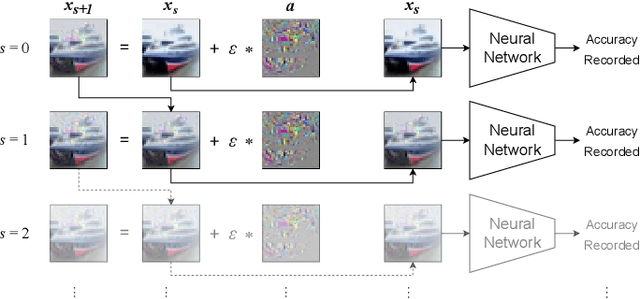

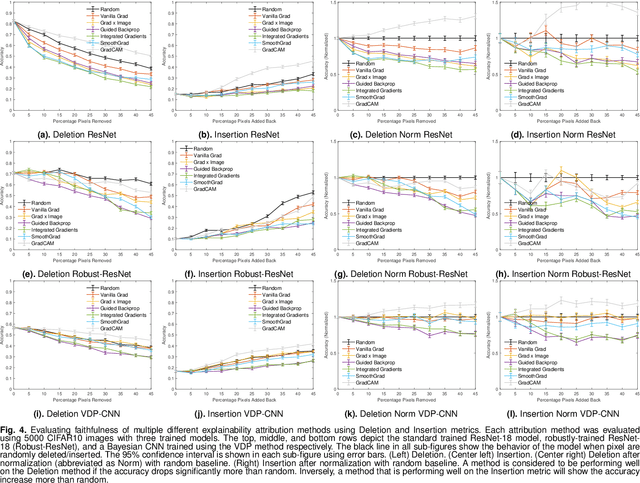
Abstract:The expansion of explainable artificial intelligence as a field of research has generated numerous methods of visualizing and understanding the black box of a machine learning model. Attribution maps are generally used to highlight the parts of the input image that influence the model to make a specific decision. On the other hand, the robustness of machine learning models to natural noise and adversarial attacks is also being actively explored. This paper focuses on evaluating methods of attribution mapping to find whether robust neural networks are more explainable. We explore this problem within the application of classification for medical imaging. Explainability research is at an impasse. There are many methods of attribution mapping, but no current consensus on how to evaluate them and determine the ones that are the best. Our experiments on multiple datasets (natural and medical imaging) and various attribution methods reveal that two popular evaluation metrics, Deletion and Insertion, have inherent limitations and yield contradictory results. We propose a new explainability faithfulness metric (called EvalAttAI) that addresses the limitations of prior metrics. Using our novel evaluation, we found that Bayesian deep neural networks using the Variational Density Propagation technique were consistently more explainable when used with the best performing attribution method, the Vanilla Gradient. However, in general, various types of robust neural networks may not be more explainable, despite these models producing more visually plausible attribution maps.
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021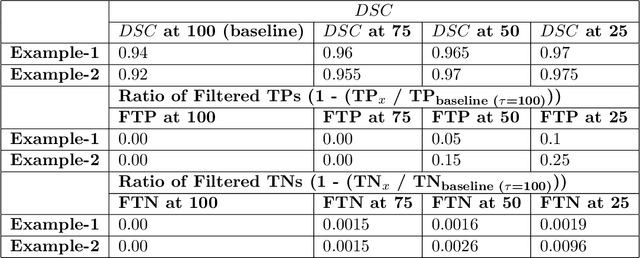
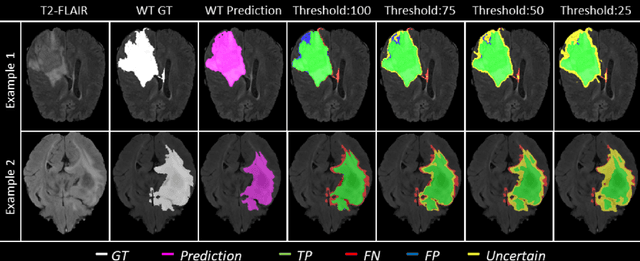
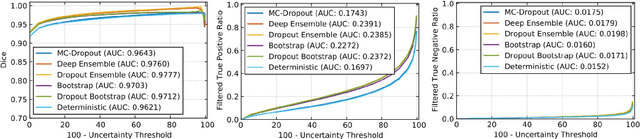
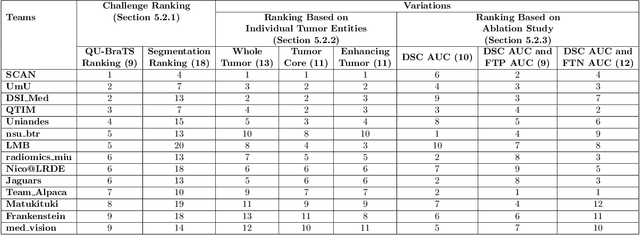
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Trustworthy Medical Segmentation with Uncertainty Estimation
Nov 30, 2021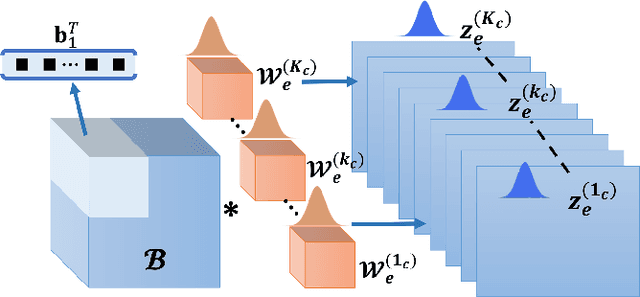
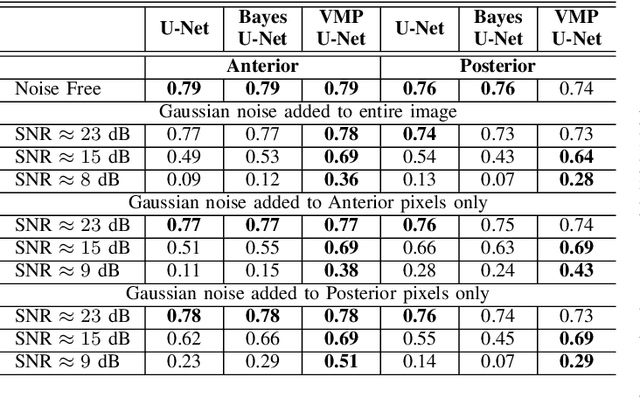
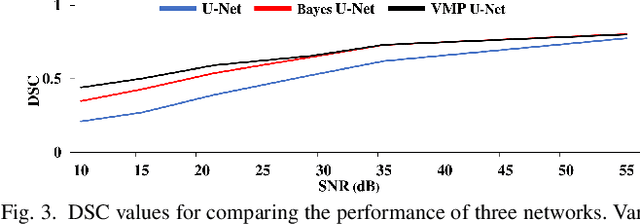
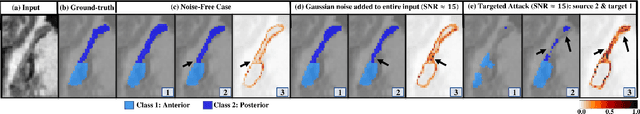
Abstract:Deep Learning (DL) holds great promise in reshaping the healthcare systems given its precision, efficiency, and objectivity. However, the brittleness of DL models to noisy and out-of-distribution inputs is ailing their deployment in the clinic. Most systems produce point estimates without further information about model uncertainty or confidence. This paper introduces a new Bayesian deep learning framework for uncertainty quantification in segmentation neural networks, specifically encoder-decoder architectures. The proposed framework uses the first-order Taylor series approximation to propagate and learn the first two moments (mean and covariance) of the distribution of the model parameters given the training data by maximizing the evidence lower bound. The output consists of two maps: the segmented image and the uncertainty map of the segmentation. The uncertainty in the segmentation decisions is captured by the covariance matrix of the predictive distribution. We evaluate the proposed framework on medical image segmentation data from Magnetic Resonances Imaging and Computed Tomography scans. Our experiments on multiple benchmark datasets demonstrate that the proposed framework is more robust to noise and adversarial attacks as compared to state-of-the-art segmentation models. Moreover, the uncertainty map of the proposed framework associates low confidence (or equivalently high uncertainty) to patches in the test input images that are corrupted with noise, artifacts or adversarial attacks. Thus, the model can self-assess its segmentation decisions when it makes an erroneous prediction or misses part of the segmentation structures, e.g., tumor, by presenting higher values in the uncertainty map.
Dilated Inception U-Net for Brain Tumor Segmentation
Aug 15, 2021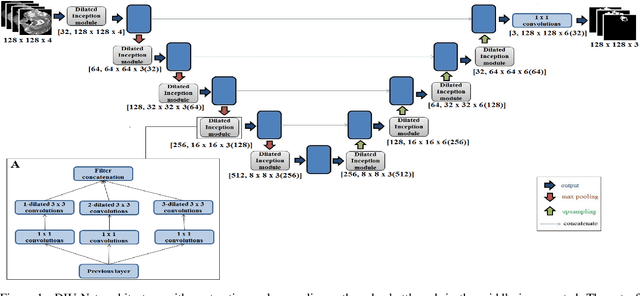
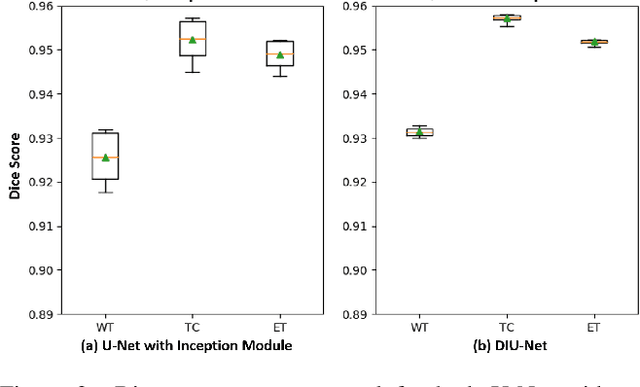
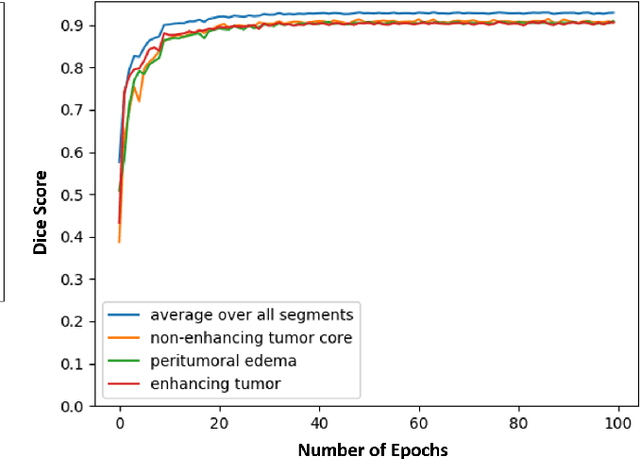
Abstract:Magnetic resonance imaging (MRI) is routinely used for brain tumor diagnosis, treatment planning, and post-treatment surveillance. Recently, various models based on deep neural networks have been proposed for the pixel-level segmentation of tumors in brain MRIs. However, the structural variations, spatial dissimilarities, and intensity inhomogeneity in MRIs make segmentation a challenging task. We propose a new end-to-end brain tumor segmentation architecture based on U-Net that integrates Inception modules and dilated convolutions into its contracting and expanding paths. This allows us to extract local structural as well as global contextual information. We performed segmentation of glioma sub-regions, including tumor core, enhancing tumor, and whole tumor using Brain Tumor Segmentation (BraTS) 2018 dataset. Our proposed model performed significantly better than the state-of-the-art U-Net-based model ($p<0.05$) for tumor core and whole tumor segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge