Ravi P. Ramachandran
SANEval: Open-Vocabulary Compositional Benchmarks with Failure-mode Diagnosis
Jan 30, 2026Abstract:The rapid progress of text-to-image (T2I) models has unlocked unprecedented creative potential, yet their ability to faithfully render complex prompts involving multiple objects, attributes, and spatial relationships remains a significant bottleneck. Progress is hampered by a lack of adequate evaluation methods; current benchmarks are often restricted to closed-set vocabularies, lack fine-grained diagnostic capabilities, and fail to provide the interpretable feedback necessary to diagnose and remedy specific compositional failures. We solve these challenges by introducing SANEval (Spatial, Attribute, and Numeracy Evaluation), a comprehensive benchmark that establishes a scalable new pipeline for open-vocabulary compositional evaluation. SANEval combines a large language model (LLM) for deep prompt understanding with an LLM-enhanced, open-vocabulary object detector to robustly evaluate compositional adherence, unconstrained by a fixed vocabulary. Through extensive experiments on six state-of-the-art T2I models, we demonstrate that SANEval's automated evaluations provide a more faithful proxy for human assessment; our metric achieves a Spearman's rank correlation with statistically different results than those of existing benchmarks across tasks of attribute binding, spatial relations, and numeracy. To facilitate future research in compositional T2I generation and evaluation, we will release the SANEval dataset and our open-source evaluation pipeline.
Future-Proofing Medical Imaging with Privacy-Preserving Federated Learning and Uncertainty Quantification: A Review
Sep 24, 2024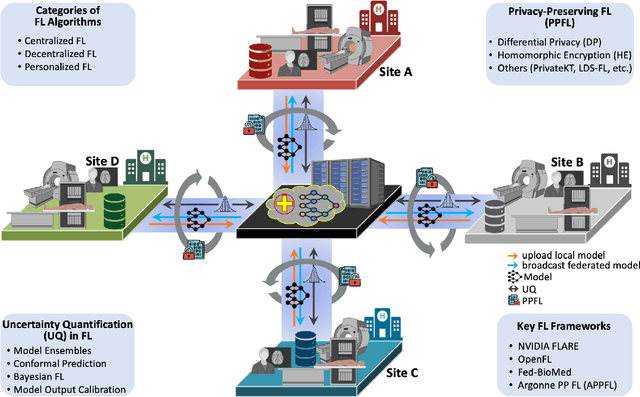
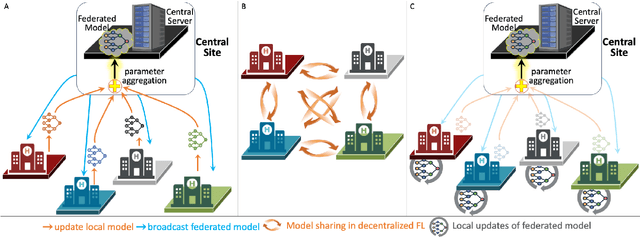
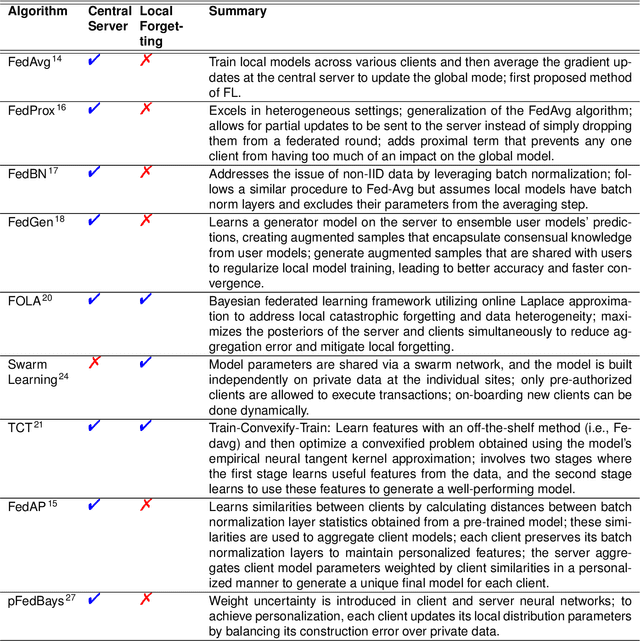
Abstract:Artificial Intelligence (AI) has demonstrated significant potential in automating various medical imaging tasks, which could soon become routine in clinical practice for disease diagnosis, prognosis, treatment planning, and post-treatment surveillance. However, the privacy concerns surrounding patient data present a major barrier to the widespread adoption of AI in medical imaging, as large, diverse training datasets are essential for developing accurate, generalizable, and robust Artificial intelligence models. Federated Learning (FL) offers a solution that enables organizations to train AI models collaboratively without sharing sensitive data. federated learning exchanges model training information, such as gradients, between the participating sites. Despite its promise, federated learning is still in its developmental stages and faces several challenges. Notably, sensitive information can still be inferred from the gradients shared during model training. Quantifying AI models' uncertainty is vital due to potential data distribution shifts post-deployment, which can affect model performance. Uncertainty quantification (UQ) in FL is particularly challenging due to data heterogeneity across participating sites. This review provides a comprehensive examination of FL, privacy-preserving FL (PPFL), and UQ in FL. We identify key gaps in current FL methodologies and propose future research directions to enhance data privacy and trustworthiness in medical imaging applications.
Privacy Preserving Federated Learning in Medical Imaging with Uncertainty Estimation
Jun 18, 2024

Abstract:Machine learning (ML) and Artificial Intelligence (AI) have fueled remarkable advancements, particularly in healthcare. Within medical imaging, ML models hold the promise of improving disease diagnoses, treatment planning, and post-treatment monitoring. Various computer vision tasks like image classification, object detection, and image segmentation are poised to become routine in clinical analysis. However, privacy concerns surrounding patient data hinder the assembly of large training datasets needed for developing and training accurate, robust, and generalizable models. Federated Learning (FL) emerges as a compelling solution, enabling organizations to collaborate on ML model training by sharing model training information (gradients) rather than data (e.g., medical images). FL's distributed learning framework facilitates inter-institutional collaboration while preserving patient privacy. However, FL, while robust in privacy preservation, faces several challenges. Sensitive information can still be gleaned from shared gradients that are passed on between organizations during model training. Additionally, in medical imaging, quantifying model confidence\uncertainty accurately is crucial due to the noise and artifacts present in the data. Uncertainty estimation in FL encounters unique hurdles due to data heterogeneity across organizations. This paper offers a comprehensive review of FL, privacy preservation, and uncertainty estimation, with a focus on medical imaging. Alongside a survey of current research, we identify gaps in the field and suggest future directions for FL research to enhance privacy and address noisy medical imaging data challenges.
Deployment of a Robust and Explainable Mortality Prediction Model: The COVID-19 Pandemic and Beyond
Nov 28, 2023Abstract:This study investigated the performance, explainability, and robustness of deployed artificial intelligence (AI) models in predicting mortality during the COVID-19 pandemic and beyond. The first study of its kind, we found that Bayesian Neural Networks (BNNs) and intelligent training techniques allowed our models to maintain performance amidst significant data shifts. Our results emphasize the importance of developing robust AI models capable of matching or surpassing clinician predictions, even under challenging conditions. Our exploration of model explainability revealed that stochastic models generate more diverse and personalized explanations thereby highlighting the need for AI models that provide detailed and individualized insights in real-world clinical settings. Furthermore, we underscored the importance of quantifying uncertainty in AI models which enables clinicians to make better-informed decisions based on reliable predictions. Our study advocates for prioritizing implementation science in AI research for healthcare and ensuring that AI solutions are practical, beneficial, and sustainable in real-world clinical environments. By addressing unique challenges and complexities in healthcare settings, researchers can develop AI models that effectively improve clinical practice and patient outcomes.
Targeted Background Removal Creates Interpretable Feature Visualizations
Jun 22, 2023



Abstract:Feature visualization is used to visualize learned features for black box machine learning models. Our approach explores an altered training process to improve interpretability of the visualizations. We argue that by using background removal techniques as a form of robust training, a network is forced to learn more human recognizable features, namely, by focusing on the main object of interest without any distractions from the background. Four different training methods were used to verify this hypothesis. The first used unmodified pictures. The second used a black background. The third utilized Gaussian noise as the background. The fourth approach employed a mix of background removed images and unmodified images. The feature visualization results show that the background removed images reveal a significant improvement over the baseline model. These new results displayed easily recognizable features from their respective classes, unlike the model trained on unmodified data.
Revisiting the Fragility of Influence Functions
Mar 22, 2023



Abstract:In the last few years, many works have tried to explain the predictions of deep learning models. Few methods, however, have been proposed to verify the accuracy or faithfulness of these explanations. Recently, influence functions, which is a method that approximates the effect that leave-one-out training has on the loss function, has been shown to be fragile. The proposed reason for their fragility remains unclear. Although previous work suggests the use of regularization to increase robustness, this does not hold in all cases. In this work, we seek to investigate the experiments performed in the prior work in an effort to understand the underlying mechanisms of influence function fragility. First, we verify influence functions using procedures from the literature under conditions where the convexity assumptions of influence functions are met. Then, we relax these assumptions and study the effects of non-convexity by using deeper models and more complex datasets. Here, we analyze the key metrics and procedures that are used to validate influence functions. Our results indicate that the validation procedures may cause the observed fragility.
EvalAttAI: A Holistic Approach to Evaluating Attribution Maps in Robust and Non-Robust Models
Mar 15, 2023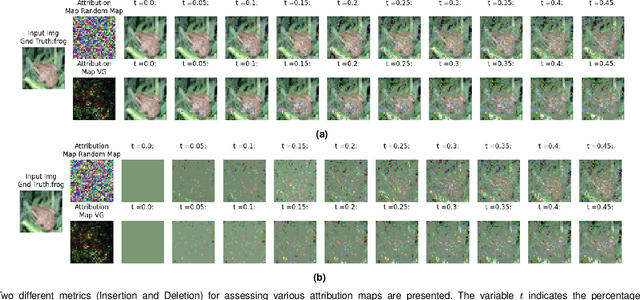
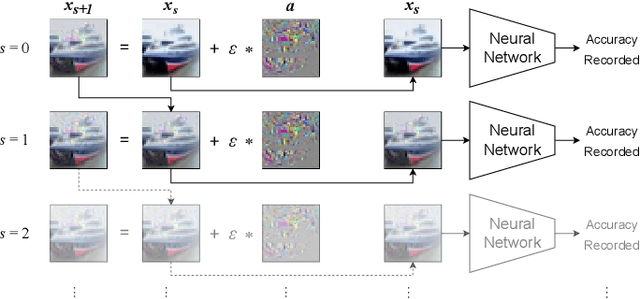

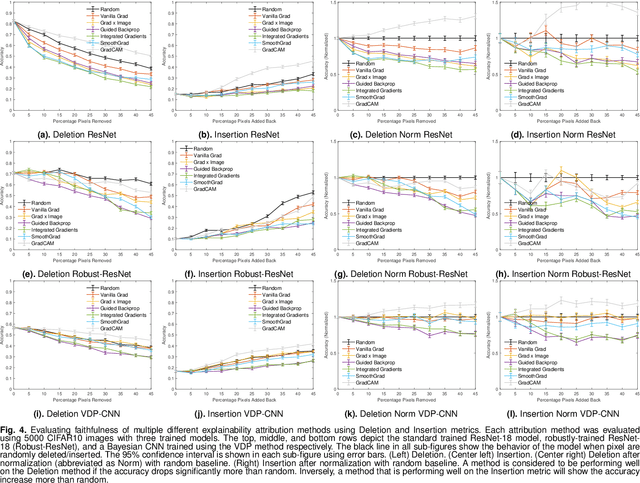
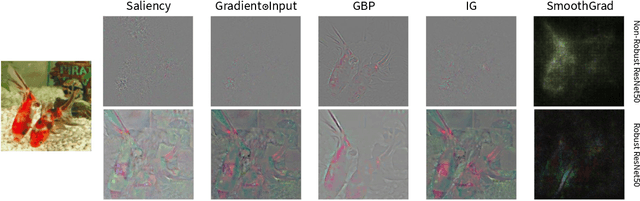
Abstract:The expansion of explainable artificial intelligence as a field of research has generated numerous methods of visualizing and understanding the black box of a machine learning model. Attribution maps are generally used to highlight the parts of the input image that influence the model to make a specific decision. On the other hand, the robustness of machine learning models to natural noise and adversarial attacks is also being actively explored. This paper focuses on evaluating methods of attribution mapping to find whether robust neural networks are more explainable. We explore this problem within the application of classification for medical imaging. Explainability research is at an impasse. There are many methods of attribution mapping, but no current consensus on how to evaluate them and determine the ones that are the best. Our experiments on multiple datasets (natural and medical imaging) and various attribution methods reveal that two popular evaluation metrics, Deletion and Insertion, have inherent limitations and yield contradictory results. We propose a new explainability faithfulness metric (called EvalAttAI) that addresses the limitations of prior metrics. Using our novel evaluation, we found that Bayesian deep neural networks using the Variational Density Propagation technique were consistently more explainable when used with the best performing attribution method, the Vanilla Gradient. However, in general, various types of robust neural networks may not be more explainable, despite these models producing more visually plausible attribution maps.
Multimodal Data Integration for Oncology in the Era of Deep Neural Networks: A Review
Mar 11, 2023Abstract:Cancer has relational information residing at varying scales, modalities, and resolutions of the acquired data, such as radiology, pathology, genomics, proteomics, and clinical records. Integrating diverse data types can improve the accuracy and reliability of cancer diagnosis and treatment. There can be disease-related information that is too subtle for humans or existing technological tools to discern visually. Traditional methods typically focus on partial or unimodal information about biological systems at individual scales and fail to encapsulate the complete spectrum of the heterogeneous nature of data. Deep neural networks have facilitated the development of sophisticated multimodal data fusion approaches that can extract and integrate relevant information from multiple sources. Recent deep learning frameworks such as Graph Neural Networks (GNNs) and Transformers have shown remarkable success in multimodal learning. This review article provides an in-depth analysis of the state-of-the-art in GNNs and Transformers for multimodal data fusion in oncology settings, highlighting notable research studies and their findings. We also discuss the foundations of multimodal learning, inherent challenges, and opportunities for integrative learning in oncology. By examining the current state and potential future developments of multimodal data integration in oncology, we aim to demonstrate the promising role that multimodal neural networks can play in cancer prevention, early detection, and treatment through informed oncology practices in personalized settings.
Transformers in Time-series Analysis: A Tutorial
Apr 28, 2022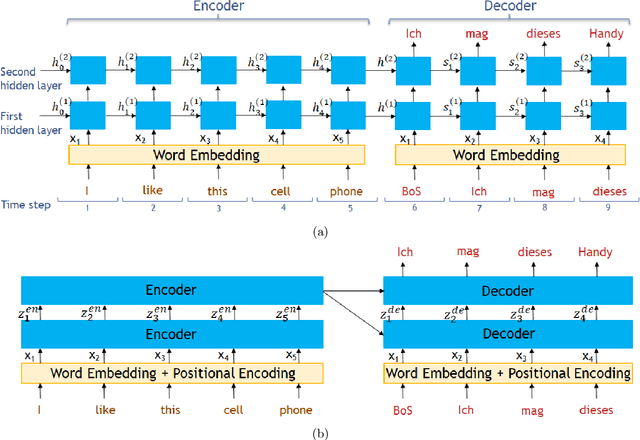
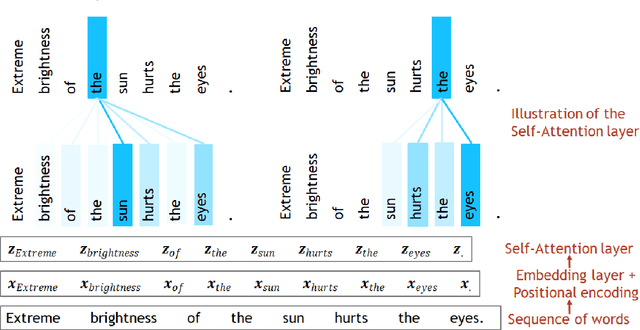
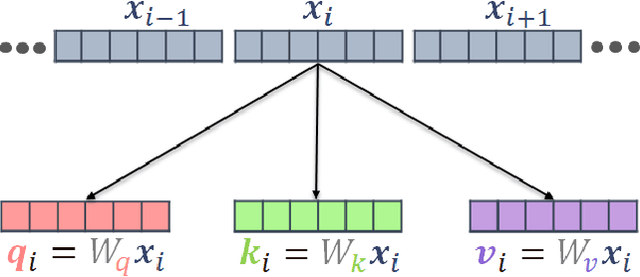
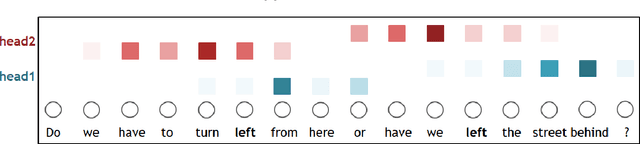
Abstract:Transformer architecture has widespread applications, particularly in Natural Language Processing and computer vision. Recently Transformers have been employed in various aspects of time-series analysis. This tutorial provides an overview of the Transformer architecture, its applications, and a collection of examples from recent research papers in time-series analysis. We delve into an explanation of the core components of the Transformer, including the self-attention mechanism, positional encoding, multi-head, and encoder/decoder. Several enhancements to the initial, Transformer architecture are highlighted to tackle time-series tasks. The tutorial also provides best practices and techniques to overcome the challenge of effectively training Transformers for time-series analysis.
Robust Explainability: A Tutorial on Gradient-Based Attribution Methods for Deep Neural Networks
Jul 28, 2021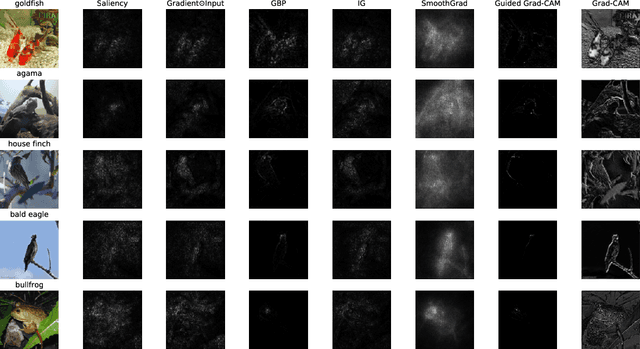
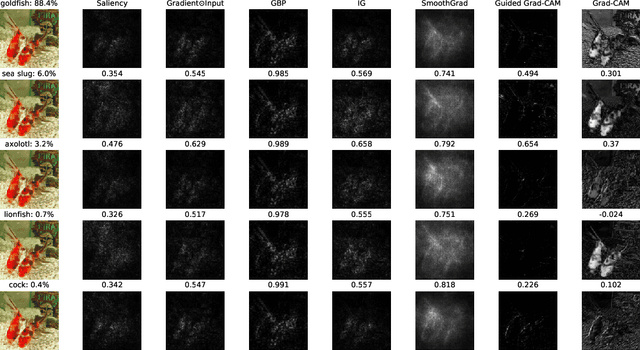
Abstract:With the rise of deep neural networks, the challenge of explaining the predictions of these networks has become increasingly recognized. While many methods for explaining the decisions of deep neural networks exist, there is currently no consensus on how to evaluate them. On the other hand, robustness is a popular topic for deep learning research; however, it is hardly talked about in explainability until very recently. In this tutorial paper, we start by presenting gradient-based interpretability methods. These techniques use gradient signals to assign the burden of the decision on the input features. Later, we discuss how gradient-based methods can be evaluated for their robustness and the role that adversarial robustness plays in having meaningful explanations. We also discuss the limitations of gradient-based methods. Finally, we present the best practices and attributes that should be examined before choosing an explainability method. We conclude with the future directions for research in the area at the convergence of robustness and explainability.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge