Asim Waqas
Rowan University
Future-Proofing Medical Imaging with Privacy-Preserving Federated Learning and Uncertainty Quantification: A Review
Sep 24, 2024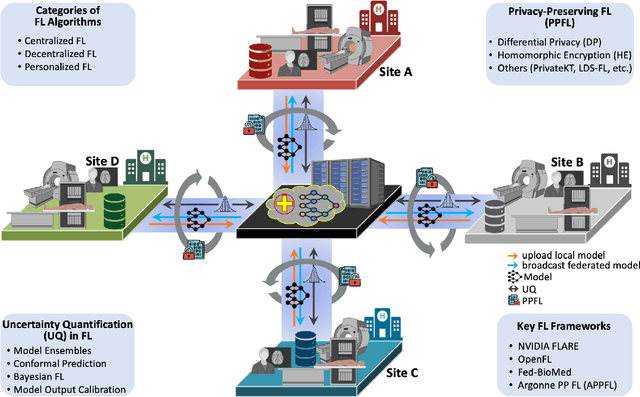
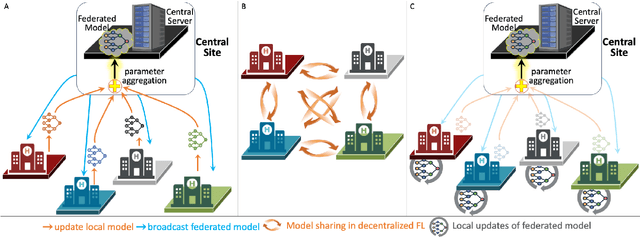
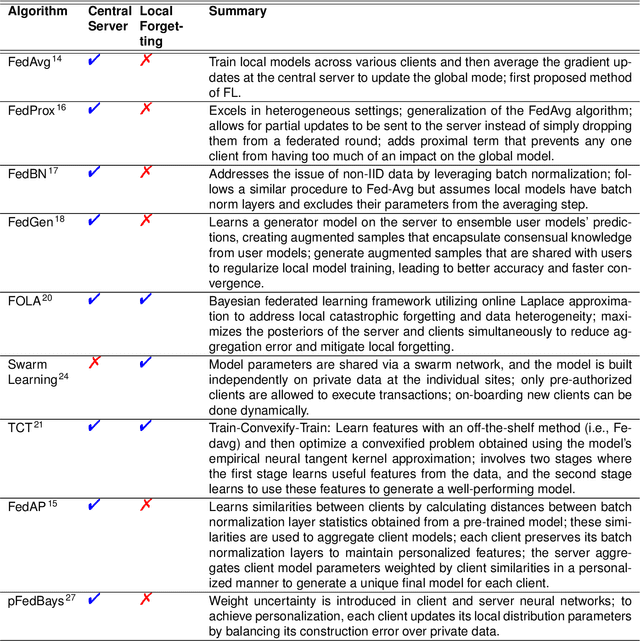
Abstract:Artificial Intelligence (AI) has demonstrated significant potential in automating various medical imaging tasks, which could soon become routine in clinical practice for disease diagnosis, prognosis, treatment planning, and post-treatment surveillance. However, the privacy concerns surrounding patient data present a major barrier to the widespread adoption of AI in medical imaging, as large, diverse training datasets are essential for developing accurate, generalizable, and robust Artificial intelligence models. Federated Learning (FL) offers a solution that enables organizations to train AI models collaboratively without sharing sensitive data. federated learning exchanges model training information, such as gradients, between the participating sites. Despite its promise, federated learning is still in its developmental stages and faces several challenges. Notably, sensitive information can still be inferred from the gradients shared during model training. Quantifying AI models' uncertainty is vital due to potential data distribution shifts post-deployment, which can affect model performance. Uncertainty quantification (UQ) in FL is particularly challenging due to data heterogeneity across participating sites. This review provides a comprehensive examination of FL, privacy-preserving FL (PPFL), and UQ in FL. We identify key gaps in current FL methodologies and propose future research directions to enhance data privacy and trustworthiness in medical imaging applications.
Embedding-based Multimodal Learning on Pan-Squamous Cell Carcinomas for Improved Survival Outcomes
Jun 11, 2024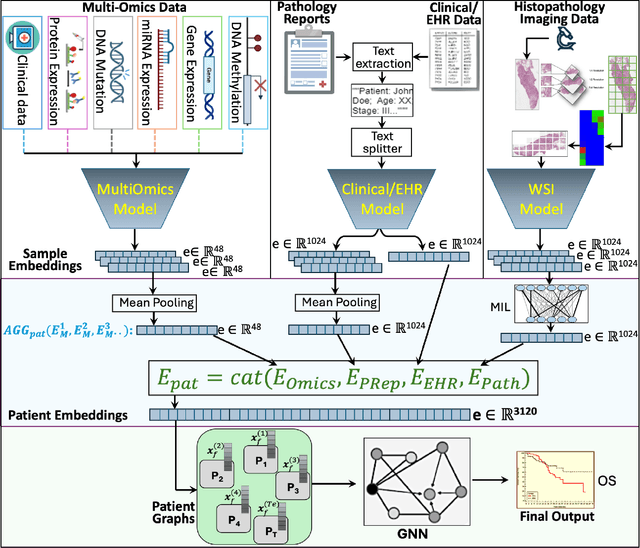
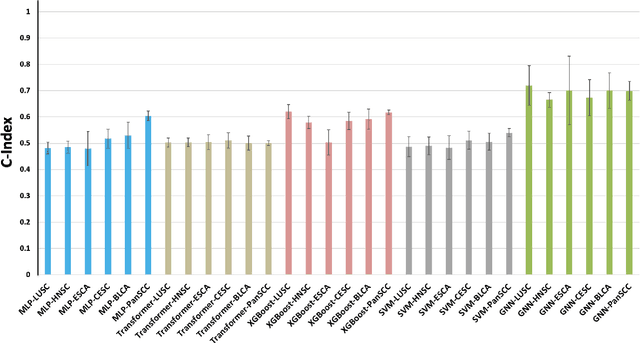

Abstract:Cancer clinics capture disease data at various scales, from genetic to organ level. Current bioinformatic methods struggle to handle the heterogeneous nature of this data, especially with missing modalities. We propose PARADIGM, a Graph Neural Network (GNN) framework that learns from multimodal, heterogeneous datasets to improve clinical outcome prediction. PARADIGM generates embeddings from multi-resolution data using foundation models, aggregates them into patient-level representations, fuses them into a unified graph, and enhances performance for tasks like survival analysis. We train GNNs on pan-Squamous Cell Carcinomas and validate our approach on Moffitt Cancer Center lung SCC data. Multimodal GNN outperforms other models in patient survival prediction. Converging individual data modalities across varying scales provides a more insightful disease view. Our solution aims to understand the patient's circumstances comprehensively, offering insights on heterogeneous data integration and the benefits of converging maximum data views.
SeNMo: A Self-Normalizing Deep Learning Model for Enhanced Multi-Omics Data Analysis in Oncology
May 13, 2024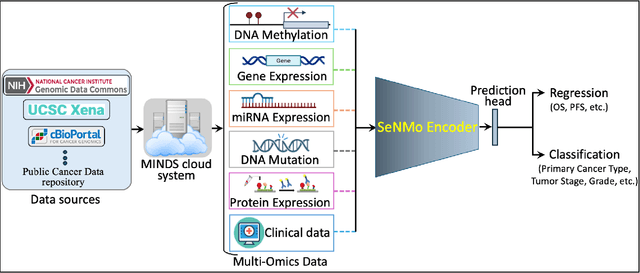
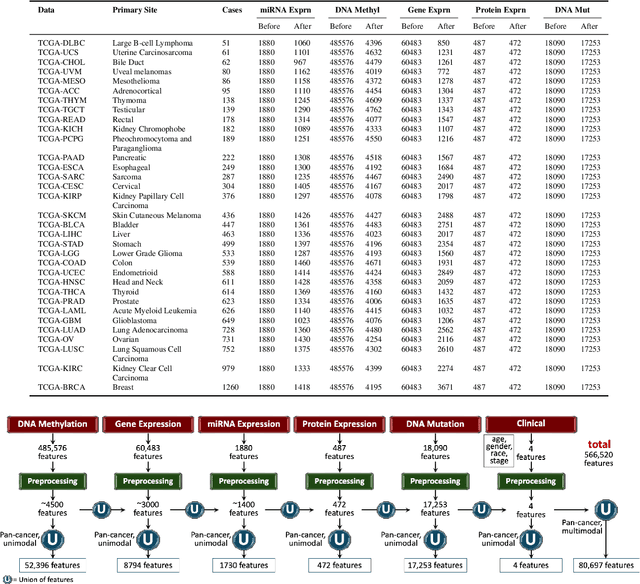


Abstract:Multi-omics research has enhanced our understanding of cancer heterogeneity and progression. Investigating molecular data through multi-omics approaches is crucial for unraveling the complex biological mechanisms underlying cancer, thereby enabling effective diagnosis, treatment, and prevention strategies. However, predicting patient outcomes through integration of all available multi-omics data is an under-study research direction. Here, we present SeNMo (Self-normalizing Network for Multi-omics), a deep neural network trained on multi-omics data across 33 cancer types. SeNMo is efficient in handling multi-omics data characterized by high-width (many features) and low-length (fewer samples) attributes. We trained SeNMo for the task of overall survival using pan-cancer data involving 33 cancer sites from Genomics Data Commons (GDC). The training data includes gene expression, DNA methylation, miRNA expression, DNA mutations, protein expression modalities, and clinical data. We evaluated the model's performance in predicting overall survival using concordance index (C-Index). SeNMo performed consistently well in training regime, with the validation C-Index of 0.76 on GDC's public data. In the testing regime, SeNMo performed with a C-Index of 0.758 on a held-out test set. The model showed an average accuracy of 99.8% on the task of classifying the primary cancer type on the pan-cancer test cohort. SeNMo proved to be a mini-foundation model for multi-omics oncology data because it demonstrated robust performance, and adaptability not only across molecular data types but also on the classification task of predicting the primary cancer type of patients. SeNMo can be further scaled to any cancer site and molecular data type. We believe SeNMo and similar models are poised to transform the oncology landscape, offering hope for more effective, efficient, and patient-centric cancer care.
HoneyBee: A Scalable Modular Framework for Creating Multimodal Oncology Datasets with Foundational Embedding Models
May 13, 2024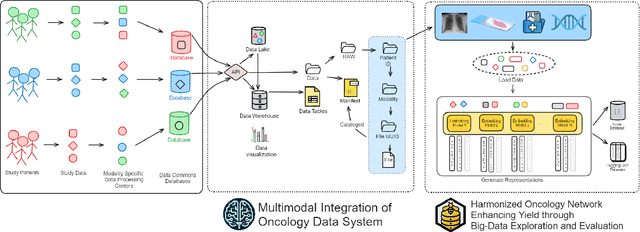
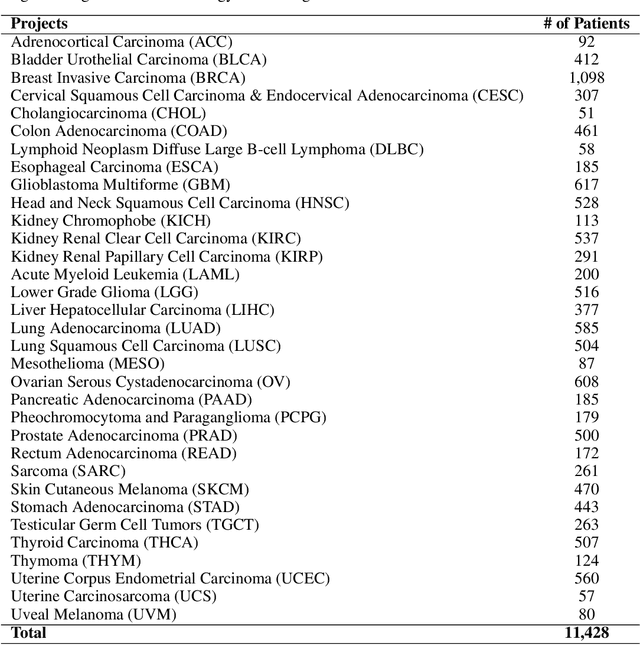

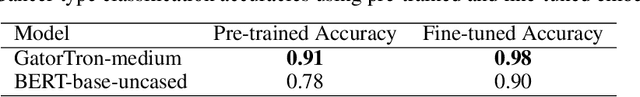
Abstract:Developing accurate machine learning models for oncology requires large-scale, high-quality multimodal datasets. However, creating such datasets remains challenging due to the complexity and heterogeneity of medical data. To address this challenge, we introduce HoneyBee, a scalable modular framework for building multimodal oncology datasets that leverages foundational models to generate representative embeddings. HoneyBee integrates various data modalities, including clinical records, imaging data, and patient outcomes. It employs data preprocessing techniques and transformer-based architectures to generate embeddings that capture the essential features and relationships within the raw medical data. The generated embeddings are stored in a structured format using Hugging Face datasets and PyTorch dataloaders for accessibility. Vector databases enable efficient querying and retrieval for machine learning applications. We demonstrate the effectiveness of HoneyBee through experiments assessing the quality and representativeness of the embeddings. The framework is designed to be extensible to other medical domains and aims to accelerate oncology research by providing high-quality, machine learning-ready datasets. HoneyBee is an ongoing open-source effort, and the code, datasets, and models are available at the project repository.
Building Flexible, Scalable, and Machine Learning-ready Multimodal Oncology Datasets
Sep 30, 2023



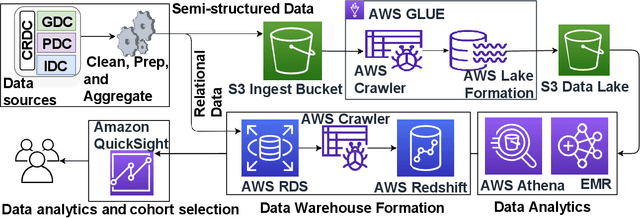
Abstract:The advancements in data acquisition, storage, and processing techniques have resulted in the rapid growth of heterogeneous medical data. Integrating radiological scans, histopathology images, and molecular information with clinical data is essential for developing a holistic understanding of the disease and optimizing treatment. The need for integrating data from multiple sources is further pronounced in complex diseases such as cancer for enabling precision medicine and personalized treatments. This work proposes Multimodal Integration of Oncology Data System (MINDS) - a flexible, scalable, and cost-effective metadata framework for efficiently fusing disparate data from public sources such as the Cancer Research Data Commons (CRDC) into an interconnected, patient-centric framework. MINDS offers an interface for exploring relationships across data types and building cohorts for developing large-scale multimodal machine learning models. By harmonizing multimodal data, MINDS aims to potentially empower researchers with greater analytical ability to uncover diagnostic and prognostic insights and enable evidence-based personalized care. MINDS tracks granular end-to-end data provenance, ensuring reproducibility and transparency. The cloud-native architecture of MINDS can handle exponential data growth in a secure, cost-optimized manner while ensuring substantial storage optimization, replication avoidance, and dynamic access capabilities. Auto-scaling, access controls, and other mechanisms guarantee pipelines' scalability and security. MINDS overcomes the limitations of existing biomedical data silos via an interoperable metadata-driven approach that represents a pivotal step toward the future of oncology data integration.
Multimodal Data Integration for Oncology in the Era of Deep Neural Networks: A Review
Mar 11, 2023Abstract:Cancer has relational information residing at varying scales, modalities, and resolutions of the acquired data, such as radiology, pathology, genomics, proteomics, and clinical records. Integrating diverse data types can improve the accuracy and reliability of cancer diagnosis and treatment. There can be disease-related information that is too subtle for humans or existing technological tools to discern visually. Traditional methods typically focus on partial or unimodal information about biological systems at individual scales and fail to encapsulate the complete spectrum of the heterogeneous nature of data. Deep neural networks have facilitated the development of sophisticated multimodal data fusion approaches that can extract and integrate relevant information from multiple sources. Recent deep learning frameworks such as Graph Neural Networks (GNNs) and Transformers have shown remarkable success in multimodal learning. This review article provides an in-depth analysis of the state-of-the-art in GNNs and Transformers for multimodal data fusion in oncology settings, highlighting notable research studies and their findings. We also discuss the foundations of multimodal learning, inherent challenges, and opportunities for integrative learning in oncology. By examining the current state and potential future developments of multimodal data integration in oncology, we aim to demonstrate the promising role that multimodal neural networks can play in cancer prevention, early detection, and treatment through informed oncology practices in personalized settings.
Exploring Robustness of Neural Networks through Graph Measures
Jun 30, 2021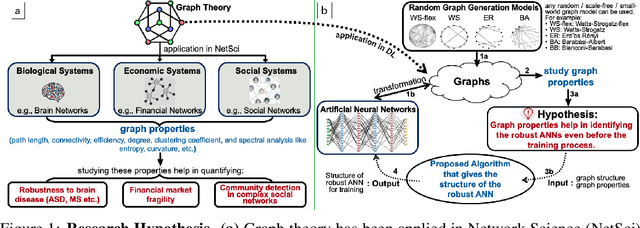
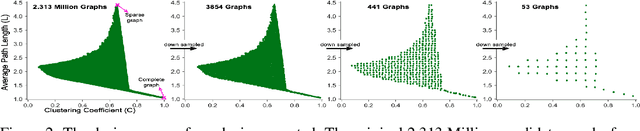
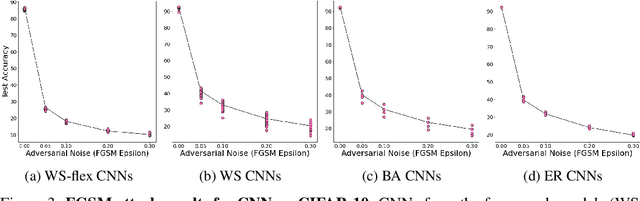
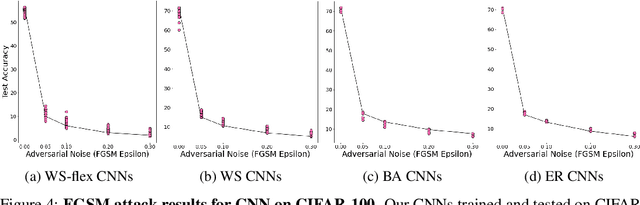
Abstract:Motivated by graph theory, artificial neural networks (ANNs) are traditionally structured as layers of neurons (nodes), which learn useful information by the passage of data through interconnections (edges). In the machine learning realm, graph structures (i.e., neurons and connections) of ANNs have recently been explored using various graph-theoretic measures linked to their predictive performance. On the other hand, in network science (NetSci), certain graph measures including entropy and curvature are known to provide insight into the robustness and fragility of real-world networks. In this work, we use these graph measures to explore the robustness of various ANNs to adversarial attacks. To this end, we (1) explore the design space of inter-layer and intra-layers connectivity regimes of ANNs in the graph domain and record their predictive performance after training under different types of adversarial attacks, (2) use graph representations for both inter-layer and intra-layers connectivity regimes to calculate various graph-theoretic measures, including curvature and entropy, and (3) analyze the relationship between these graph measures and the adversarial performance of ANNs. We show that curvature and entropy, while operating in the graph domain, can quantify the robustness of ANNs without having to train these ANNs. Our results suggest that the real-world networks, including brain networks, financial networks, and social networks may provide important clues to the neural architecture search for robust ANNs. We propose a search strategy that efficiently finds robust ANNs amongst a set of well-performing ANNs without having a need to train all of these ANNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge