Hamza Farooq
University of Minnesota
UrduBench: An Urdu Reasoning Benchmark using Contextually Ensembled Translations with Human-in-the-Loop
Jan 28, 2026Abstract:Recent advances in large language models (LLMs) have led to strong reasoning capabilities; however, evaluating such models in low-resource languages remains challenging due to the lack of standardized benchmarks. In particular, Urdu reasoning evaluation has been limited by the sensitivity of machine translation and an emphasis on general language tasks rather than reasoning benchmarks. In this paper, we propose a contextually ensembled translation framework with human-in-the-loop validation that leverages multiple translation systems to develop Urdu reasoning benchmarks while preserving contextual and structural integrity. Using this framework, we translate widely adopted reasoning and question-answering benchmarks, including MGSM, MATH-500, CommonSenseQA, and OpenBookQA, into Urdu, collectively referred to as UrduBench, and conduct a comprehensive evaluation of both reasoning-oriented and instruction-tuned LLMs across multiple prompting strategies. Our analysis reveals performance differences across (1) four datasets, (2) five task difficulty levels, (3) diverse model architectures, (4) multiple model scaling settings, and (5) language consistency tests. We find that multi-step and symbolic reasoning tasks pose significant challenges in Urdu, and that stable language alignment is a critical prerequisite for robust reasoning. Overall, our work establishes a scalable methodology for standardized reasoning evaluation in Urdu and provides empirical insights into multilingual reasoning failures. This experimental setup is also broadly applicable to other low-resource languages. The code and datasets will be publicly released.
Query Attribute Modeling: Improving search relevance with Semantic Search and Meta Data Filtering
Aug 06, 2025Abstract:This study introduces Query Attribute Modeling (QAM), a hybrid framework that enhances search precision and relevance by decomposing open text queries into structured metadata tags and semantic elements. QAM addresses traditional search limitations by automatically extracting metadata filters from free-form text queries, reducing noise and enabling focused retrieval of relevant items. Experimental evaluation using the Amazon Toys Reviews dataset (10,000 unique items with 40,000+ reviews and detailed product attributes) demonstrated QAM's superior performance, achieving a mean average precision at 5 (mAP@5) of 52.99\%. This represents significant improvement over conventional methods, including BM25 keyword search, encoder-based semantic similarity search, cross-encoder re-ranking, and hybrid search combining BM25 and semantic results via Reciprocal Rank Fusion (RRF). The results establish QAM as a robust solution for Enterprise Search applications, particularly in e-commerce systems.
Robust and Fine-Grained Detection of AI Generated Texts
Apr 16, 2025Abstract:An ideal detection system for machine generated content is supposed to work well on any generator as many more advanced LLMs come into existence day by day. Existing systems often struggle with accurately identifying AI-generated content over shorter texts. Further, not all texts might be entirely authored by a human or LLM, hence we focused more over partial cases i.e human-LLM co-authored texts. Our paper introduces a set of models built for the task of token classification which are trained on an extensive collection of human-machine co-authored texts, which performed well over texts of unseen domains, unseen generators, texts by non-native speakers and those with adversarial inputs. We also introduce a new dataset of over 2.4M such texts mostly co-authored by several popular proprietary LLMs over 23 languages. We also present findings of our models' performance over each texts of each domain and generator. Additional findings include comparison of performance against each adversarial method, length of input texts and characteristics of generated texts compared to the original human authored texts.
Improving Multilingual Capabilities with Cultural and Local Knowledge in Large Language Models While Enhancing Native Performance
Apr 13, 2025



Abstract:Large Language Models (LLMs) have shown remarkable capabilities, but their development has primarily focused on English and other high-resource languages, leaving many languages underserved. We present our latest Hindi-English bi-lingual LLM \textbf{Mantra-14B} with ~3\% average improvement in benchmark scores over both languages, outperforming models twice its size. Using a curated dataset composed of English and Hindi instruction data of 485K samples, we instruction tuned models such as Qwen-2.5-14B-Instruct and Phi-4 to improve performance over both English and Hindi. Our experiments encompassing seven different LLMs of varying parameter sizes and over 140 training attempts with varying English-Hindi training data ratios demonstrated that it is possible to significantly improve multilingual performance without compromising native performance. Further, our approach avoids resource-intensive techniques like vocabulary expansion or architectural modifications, thus keeping the model size small. Our results indicate that modest fine-tuning with culturally and locally informed data can bridge performance gaps without incurring significant computational overhead. We release our training code, datasets, and models under mit and apache licenses to aid further research towards under-represented and low-resource languages.
LSSF-Net: Lightweight Segmentation with Self-Awareness, Spatial Attention, and Focal Modulation
Sep 03, 2024

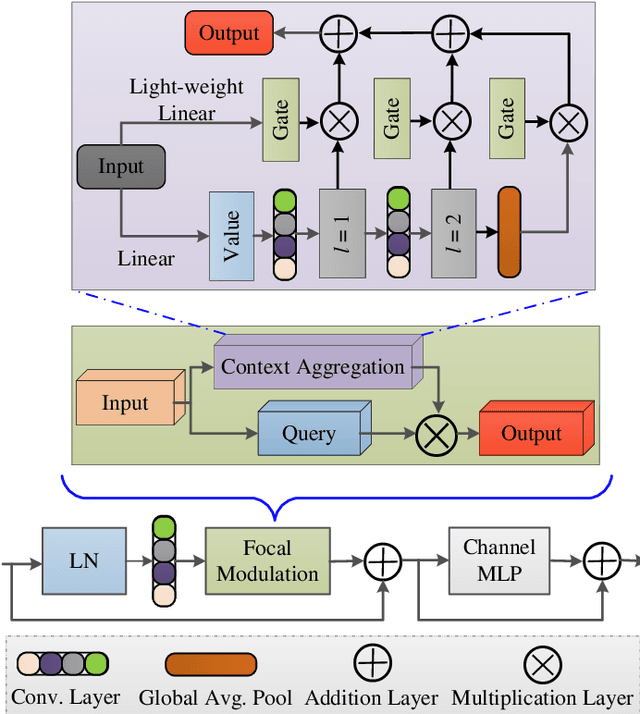
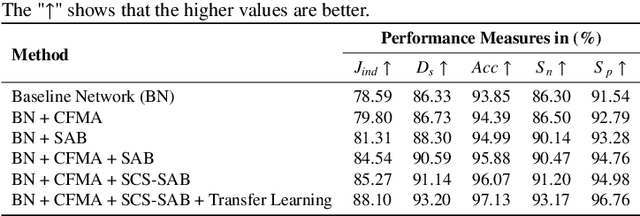
Abstract:Accurate segmentation of skin lesions within dermoscopic images plays a crucial role in the timely identification of skin cancer for computer-aided diagnosis on mobile platforms. However, varying shapes of the lesions, lack of defined edges, and the presence of obstructions such as hair strands and marker colors make this challenge more complex. \textcolor{red}Additionally, skin lesions often exhibit subtle variations in texture and color that are difficult to differentiate from surrounding healthy skin, necessitating models that can capture both fine-grained details and broader contextual information. Currently, melanoma segmentation models are commonly based on fully connected networks and U-Nets. However, these models often struggle with capturing the complex and varied characteristics of skin lesions, such as the presence of indistinct boundaries and diverse lesion appearances, which can lead to suboptimal segmentation performance.To address these challenges, we propose a novel lightweight network specifically designed for skin lesion segmentation utilizing mobile devices, featuring a minimal number of learnable parameters (only 0.8 million). This network comprises an encoder-decoder architecture that incorporates conformer-based focal modulation attention, self-aware local and global spatial attention, and split channel-shuffle. The efficacy of our model has been evaluated on four well-established benchmark datasets for skin lesion segmentation: ISIC 2016, ISIC 2017, ISIC 2018, and PH2. Empirical findings substantiate its state-of-the-art performance, notably reflected in a high Jaccard index.
SeNMo: A Self-Normalizing Deep Learning Model for Enhanced Multi-Omics Data Analysis in Oncology
May 13, 2024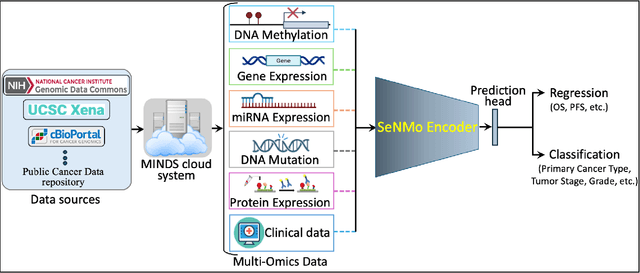
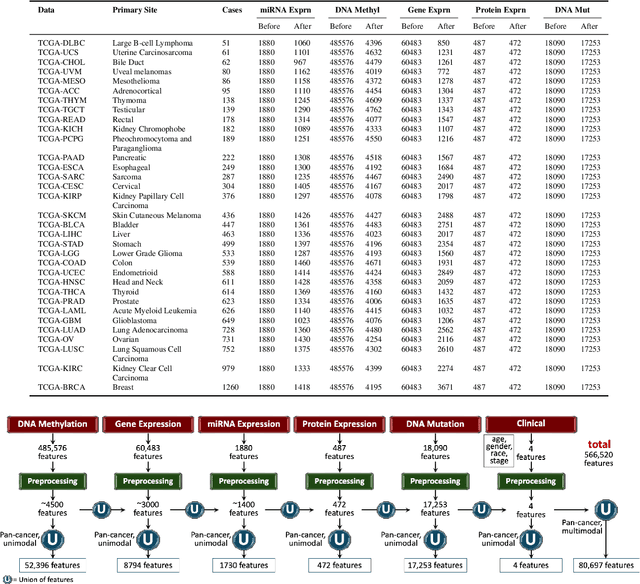


Abstract:Multi-omics research has enhanced our understanding of cancer heterogeneity and progression. Investigating molecular data through multi-omics approaches is crucial for unraveling the complex biological mechanisms underlying cancer, thereby enabling effective diagnosis, treatment, and prevention strategies. However, predicting patient outcomes through integration of all available multi-omics data is an under-study research direction. Here, we present SeNMo (Self-normalizing Network for Multi-omics), a deep neural network trained on multi-omics data across 33 cancer types. SeNMo is efficient in handling multi-omics data characterized by high-width (many features) and low-length (fewer samples) attributes. We trained SeNMo for the task of overall survival using pan-cancer data involving 33 cancer sites from Genomics Data Commons (GDC). The training data includes gene expression, DNA methylation, miRNA expression, DNA mutations, protein expression modalities, and clinical data. We evaluated the model's performance in predicting overall survival using concordance index (C-Index). SeNMo performed consistently well in training regime, with the validation C-Index of 0.76 on GDC's public data. In the testing regime, SeNMo performed with a C-Index of 0.758 on a held-out test set. The model showed an average accuracy of 99.8% on the task of classifying the primary cancer type on the pan-cancer test cohort. SeNMo proved to be a mini-foundation model for multi-omics oncology data because it demonstrated robust performance, and adaptability not only across molecular data types but also on the classification task of predicting the primary cancer type of patients. SeNMo can be further scaled to any cancer site and molecular data type. We believe SeNMo and similar models are poised to transform the oncology landscape, offering hope for more effective, efficient, and patient-centric cancer care.
Exploring Robustness of Neural Networks through Graph Measures
Jun 30, 2021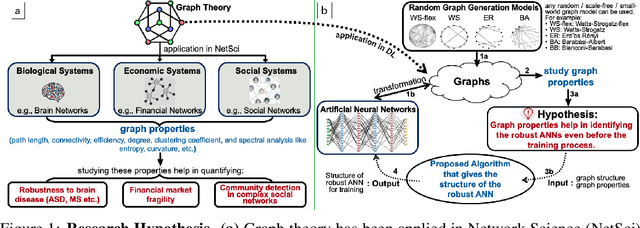
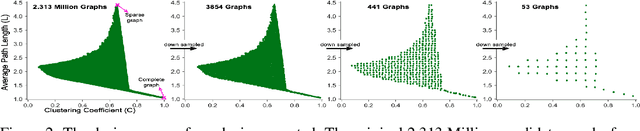
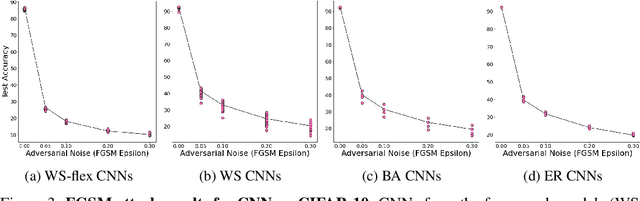
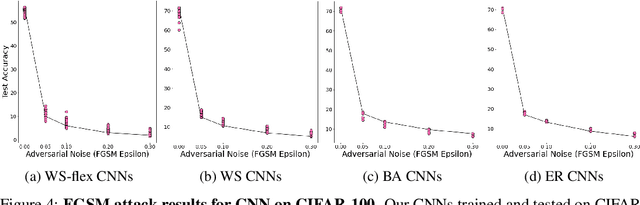
Abstract:Motivated by graph theory, artificial neural networks (ANNs) are traditionally structured as layers of neurons (nodes), which learn useful information by the passage of data through interconnections (edges). In the machine learning realm, graph structures (i.e., neurons and connections) of ANNs have recently been explored using various graph-theoretic measures linked to their predictive performance. On the other hand, in network science (NetSci), certain graph measures including entropy and curvature are known to provide insight into the robustness and fragility of real-world networks. In this work, we use these graph measures to explore the robustness of various ANNs to adversarial attacks. To this end, we (1) explore the design space of inter-layer and intra-layers connectivity regimes of ANNs in the graph domain and record their predictive performance after training under different types of adversarial attacks, (2) use graph representations for both inter-layer and intra-layers connectivity regimes to calculate various graph-theoretic measures, including curvature and entropy, and (3) analyze the relationship between these graph measures and the adversarial performance of ANNs. We show that curvature and entropy, while operating in the graph domain, can quantify the robustness of ANNs without having to train these ANNs. Our results suggest that the real-world networks, including brain networks, financial networks, and social networks may provide important clues to the neural architecture search for robust ANNs. We propose a search strategy that efficiently finds robust ANNs amongst a set of well-performing ANNs without having a need to train all of these ANNs.
Fitting IVIM with Variable Projection and Simplicial Optimization
Oct 07, 2019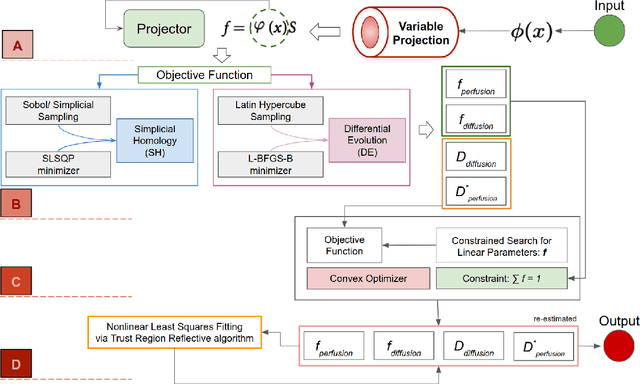
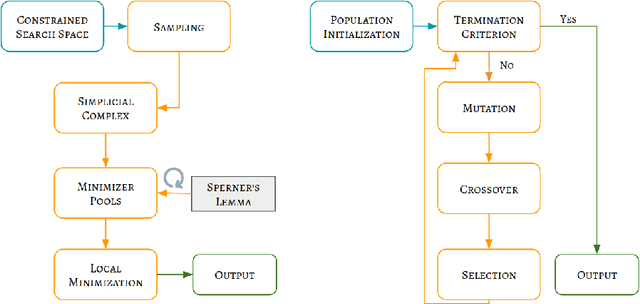
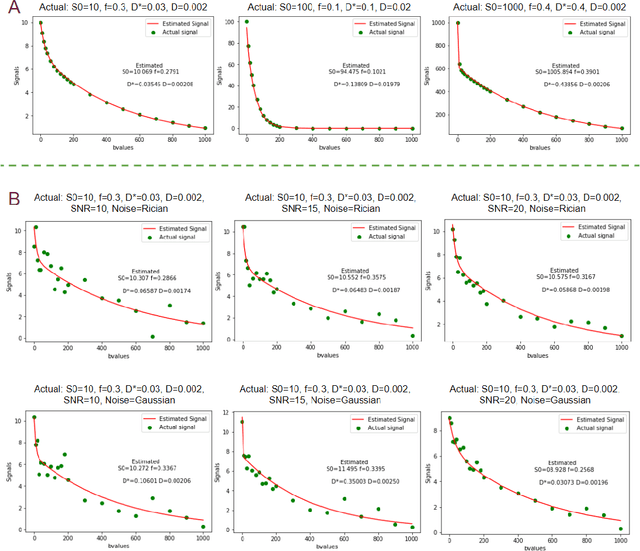
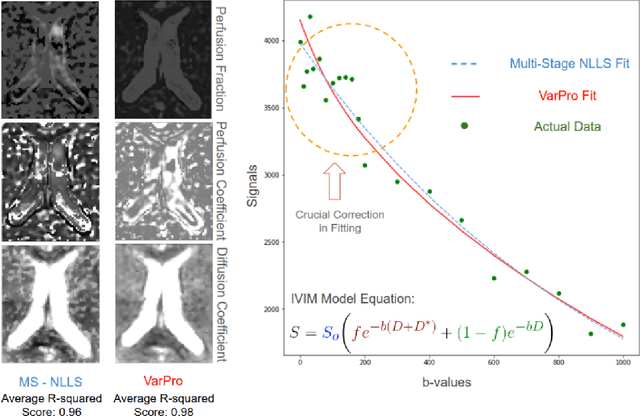
Abstract:Fitting multi-exponential models to Diffusion MRI (dMRI) data has always been challenging due to various underlying complexities. In this work, we introduce a novel and robust fitting framework for the standard two-compartment IVIM microstructural model. This framework provides a significant improvement over the existing methods and helps estimate the associated diffusion and perfusion parameters of IVIM in an automatic manner. As a part of this work we provide capabilities to switch between more advanced global optimization methods such as simplicial homology (SH) and differential evolution (DE). Our experiments show that the results obtained from this simultaneous fitting procedure disentangle the model parameters in a reduced subspace. The proposed framework extends the seminal work originated in the MIX framework, with improved procedures for multi-stage fitting. This framework has been made available as an open-source Python implementation and disseminated to the community through the DIPY project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge