Fergus Imrie
Assessing the Chemical Intelligence of Large Language Models
May 12, 2025Abstract:Large Language Models are versatile, general-purpose tools with a wide range of applications. Recently, the advent of "reasoning models" has led to substantial improvements in their abilities in advanced problem-solving domains such as mathematics and software engineering. In this work, we assessed the ability of reasoning models to directly perform chemistry tasks, without any assistance from external tools. We created a novel benchmark, called ChemIQ, which consists of 796 questions assessing core concepts in organic chemistry, focused on molecular comprehension and chemical reasoning. Unlike previous benchmarks, which primarily use multiple choice formats, our approach requires models to construct short-answer responses, more closely reflecting real-world applications. The reasoning models, exemplified by OpenAI's o3-mini, correctly answered 28%-59% of questions depending on the reasoning level used, with higher reasoning levels significantly increasing performance on all tasks. These models substantially outperformed the non-reasoning model, GPT-4o, which achieved only 7% accuracy. We found that Large Language Models can now convert SMILES strings to IUPAC names, a task earlier models were unable to perform. Additionally, we show that the latest reasoning models can elucidate structures from 1H and 13C NMR data, correctly generating SMILES strings for 74% of molecules containing up to 10 heavy atoms, and in one case solving a structure comprising 21 heavy atoms. For each task, we found evidence that the reasoning process mirrors that of a human chemist. Our results demonstrate that the latest reasoning models have the ability to perform advanced chemical reasoning.
Automated Ensemble Multimodal Machine Learning for Healthcare
Jul 25, 2024



Abstract:The application of machine learning in medicine and healthcare has led to the creation of numerous diagnostic and prognostic models. However, despite their success, current approaches generally issue predictions using data from a single modality. This stands in stark contrast with clinician decision-making which employs diverse information from multiple sources. While several multimodal machine learning approaches exist, significant challenges in developing multimodal systems remain that are hindering clinical adoption. In this paper, we introduce a multimodal framework, AutoPrognosis-M, that enables the integration of structured clinical (tabular) data and medical imaging using automated machine learning. AutoPrognosis-M incorporates 17 imaging models, including convolutional neural networks and vision transformers, and three distinct multimodal fusion strategies. In an illustrative application using a multimodal skin lesion dataset, we highlight the importance of multimodal machine learning and the power of combining multiple fusion strategies using ensemble learning. We have open-sourced our framework as a tool for the community and hope it will accelerate the uptake of multimodal machine learning in healthcare and spur further innovation.
You can't handle the truth: Data-centric insights improve pseudo-labeling
Jun 19, 2024Abstract:Pseudo-labeling is a popular semi-supervised learning technique to leverage unlabeled data when labeled samples are scarce. The generation and selection of pseudo-labels heavily rely on labeled data. Existing approaches implicitly assume that the labeled data is gold standard and 'perfect'. However, this can be violated in reality with issues such as mislabeling or ambiguity. We address this overlooked aspect and show the importance of investigating labeled data quality to improve any pseudo-labeling method. Specifically, we introduce a novel data characterization and selection framework called DIPS to extend pseudo-labeling. We select useful labeled and pseudo-labeled samples via analysis of learning dynamics. We demonstrate the applicability and impact of DIPS for various pseudo-labeling methods across an extensive range of real-world tabular and image datasets. Additionally, DIPS improves data efficiency and reduces the performance distinctions between different pseudo-labelers. Overall, we highlight the significant benefits of a data-centric rethinking of pseudo-labeling in real-world settings.
Dissecting Sample Hardness: A Fine-Grained Analysis of Hardness Characterization Methods for Data-Centric AI
Mar 07, 2024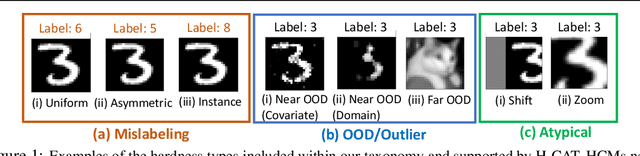
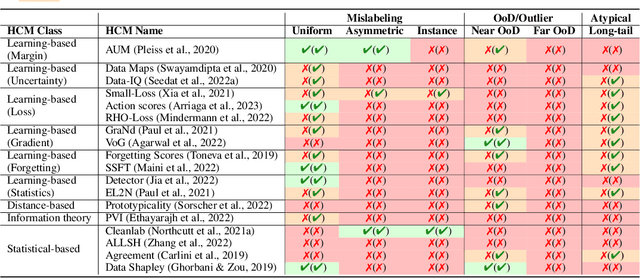
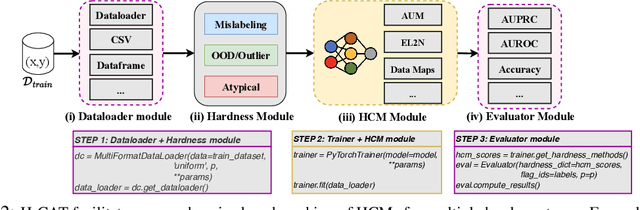
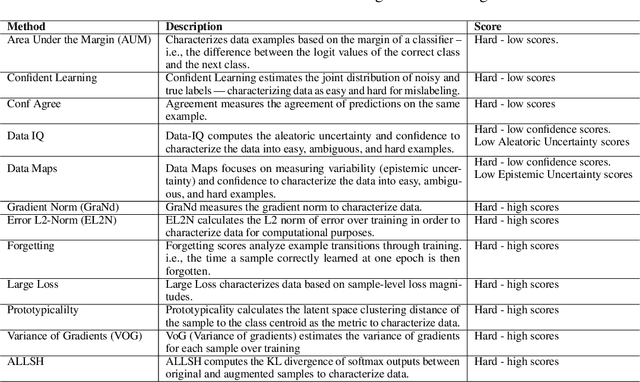
Abstract:Characterizing samples that are difficult to learn from is crucial to developing highly performant ML models. This has led to numerous Hardness Characterization Methods (HCMs) that aim to identify "hard" samples. However, there is a lack of consensus regarding the definition and evaluation of "hardness". Unfortunately, current HCMs have only been evaluated on specific types of hardness and often only qualitatively or with respect to downstream performance, overlooking the fundamental quantitative identification task. We address this gap by presenting a fine-grained taxonomy of hardness types. Additionally, we propose the Hardness Characterization Analysis Toolkit (H-CAT), which supports comprehensive and quantitative benchmarking of HCMs across the hardness taxonomy and can easily be extended to new HCMs, hardness types, and datasets. We use H-CAT to evaluate 13 different HCMs across 8 hardness types. This comprehensive evaluation encompassing over 14K setups uncovers strengths and weaknesses of different HCMs, leading to practical tips to guide HCM selection and future development. Our findings highlight the need for more comprehensive HCM evaluation, while we hope our hardness taxonomy and toolkit will advance the principled evaluation and uptake of data-centric AI methods.
A Neural Framework for Generalized Causal Sensitivity Analysis
Nov 27, 2023



Abstract:Unobserved confounding is common in many applications, making causal inference from observational data challenging. As a remedy, causal sensitivity analysis is an important tool to draw causal conclusions under unobserved confounding with mathematical guarantees. In this paper, we propose NeuralCSA, a neural framework for generalized causal sensitivity analysis. Unlike previous work, our framework is compatible with (i) a large class of sensitivity models, including the marginal sensitivity model, f-sensitivity models, and Rosenbaum's sensitivity model; (ii) different treatment types (i.e., binary and continuous); and (iii) different causal queries, including (conditional) average treatment effects and simultaneous effects on multiple outcomes. The generality of \frameworkname is achieved by learning a latent distribution shift that corresponds to a treatment intervention using two conditional normalizing flows. We provide theoretical guarantees that NeuralCSA is able to infer valid bounds on the causal query of interest and also demonstrate this empirically using both simulated and real-world data.
Can You Rely on Your Model Evaluation? Improving Model Evaluation with Synthetic Test Data
Oct 25, 2023Abstract:Evaluating the performance of machine learning models on diverse and underrepresented subgroups is essential for ensuring fairness and reliability in real-world applications. However, accurately assessing model performance becomes challenging due to two main issues: (1) a scarcity of test data, especially for small subgroups, and (2) possible distributional shifts in the model's deployment setting, which may not align with the available test data. In this work, we introduce 3S Testing, a deep generative modeling framework to facilitate model evaluation by generating synthetic test sets for small subgroups and simulating distributional shifts. Our experiments demonstrate that 3S Testing outperforms traditional baselines -- including real test data alone -- in estimating model performance on minority subgroups and under plausible distributional shifts. In addition, 3S offers intervals around its performance estimates, exhibiting superior coverage of the ground truth compared to existing approaches. Overall, these results raise the question of whether we need a paradigm shift away from limited real test data towards synthetic test data.
Redefining Digital Health Interfaces with Large Language Models
Oct 05, 2023Abstract:Digital health tools have the potential to significantly improve the delivery of healthcare services. However, their use remains comparatively limited due, in part, to challenges surrounding usability and trust. Recently, Large Language Models (LLMs) have emerged as general-purpose models with the ability to process complex information and produce human-quality text, presenting a wealth of potential applications in healthcare. Directly applying LLMs in clinical settings is not straightforward, with LLMs susceptible to providing inconsistent or nonsensical answers. We demonstrate how LLMs can utilize external tools to provide a novel interface between clinicians and digital technologies. This enhances the utility and practical impact of digital healthcare tools and AI models while addressing current issues with using LLM in clinical settings such as hallucinations. We illustrate our approach with examples from cardiovascular disease and diabetes risk prediction, highlighting the benefit compared to traditional interfaces for digital tools.
Machine Learning with Requirements: a Manifesto
Apr 07, 2023Abstract:In the recent years, machine learning has made great advancements that have been at the root of many breakthroughs in different application domains. However, it is still an open issue how make them applicable to high-stakes or safety-critical application domains, as they can often be brittle and unreliable. In this paper, we argue that requirements definition and satisfaction can go a long way to make machine learning models even more fitting to the real world, especially in critical domains. To this end, we present two problems in which (i) requirements arise naturally, (ii) machine learning models are or can be fruitfully deployed, and (iii) neglecting the requirements can have dramatic consequences. We show how the requirements specification can be fruitfully integrated into the standard machine learning development pipeline, proposing a novel pyramid development process in which requirements definition may impact all the subsequent phases in the pipeline, and viceversa.
TANGOS: Regularizing Tabular Neural Networks through Gradient Orthogonalization and Specialization
Mar 09, 2023Abstract:Despite their success with unstructured data, deep neural networks are not yet a panacea for structured tabular data. In the tabular domain, their efficiency crucially relies on various forms of regularization to prevent overfitting and provide strong generalization performance. Existing regularization techniques include broad modelling decisions such as choice of architecture, loss functions, and optimization methods. In this work, we introduce Tabular Neural Gradient Orthogonalization and Specialization (TANGOS), a novel framework for regularization in the tabular setting built on latent unit attributions. The gradient attribution of an activation with respect to a given input feature suggests how the neuron attends to that feature, and is often employed to interpret the predictions of deep networks. In TANGOS, we take a different approach and incorporate neuron attributions directly into training to encourage orthogonalization and specialization of latent attributions in a fully-connected network. Our regularizer encourages neurons to focus on sparse, non-overlapping input features and results in a set of diverse and specialized latent units. In the tabular domain, we demonstrate that our approach can lead to improved out-of-sample generalization performance, outperforming other popular regularization methods. We provide insight into why our regularizer is effective and demonstrate that TANGOS can be applied jointly with existing methods to achieve even greater generalization performance.
SurvivalGAN: Generating Time-to-Event Data for Survival Analysis
Feb 24, 2023Abstract:Synthetic data is becoming an increasingly promising technology, and successful applications can improve privacy, fairness, and data democratization. While there are many methods for generating synthetic tabular data, the task remains non-trivial and unexplored for specific scenarios. One such scenario is survival data. Here, the key difficulty is censoring: for some instances, we are not aware of the time of event, or if one even occurred. Imbalances in censoring and time horizons cause generative models to experience three new failure modes specific to survival analysis: (1) generating too few at-risk members; (2) generating too many at-risk members; and (3) censoring too early. We formalize these failure modes and provide three new generative metrics to quantify them. Following this, we propose SurvivalGAN, a generative model that handles survival data firstly by addressing the imbalance in the censoring and event horizons, and secondly by using a dedicated mechanism for approximating time-to-event/censoring. We evaluate this method via extensive experiments on medical datasets. SurvivalGAN outperforms multiple baselines at generating survival data, and in particular addresses the failure modes as measured by the new metrics, in addition to improving downstream performance of survival models trained on the synthetic data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge