Felix Hofmann
Department of Diagnostic and Interventional Radiology, School of Medicine, Klinikum rechts der Isar, Technical University of Munich, Germany
Single-exposure elemental differentiation and texture-sensitive phase-retrieval imaging with a neutron counting micro-channel plate detector
Nov 09, 2023



Abstract:Micro-channel plate (MCP) detectors, when used at pulsed-neutron-source instruments, offer the possibility of high spatial resolution and high contrast imaging with pixel-level spectroscopic information. Here we demonstrate the possibility of multimodal analysis including total neutron cross-section spectra measurements, quantitative material differentiation imaging and texture-sensitive in-line phase imaging, from a single exposure using an MCP detector. This multimodal approach operates in full-field imaging mode, with the neutron transmission spectra acquired at each individual detector pixel. Due to the polychromatic nature of the beam and spectroscopic resolving capability of the detector, no energy scanning is required. Good agreement with the library reference data is demonstrated for neutron cross-section spectra measurements. Two different images corresponding to two selected energy bandwidths are used for elemental differentiation imaging. Moreover, the presence of changes in texture, i.e., preferred grain orientation, in the sample is identified from our phase-retrieval imaging results.
Optimizing Convolutional Neural Networks for Chronic Obstructive Pulmonary Disease Detection in Clinical Computed Tomography Imaging
Mar 13, 2023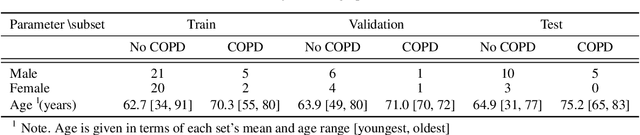
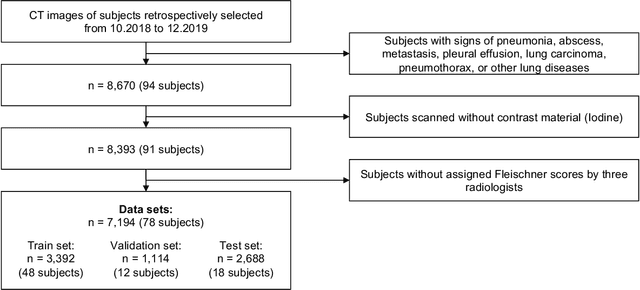
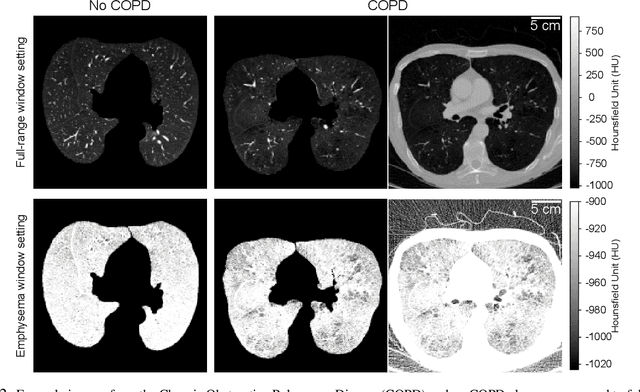
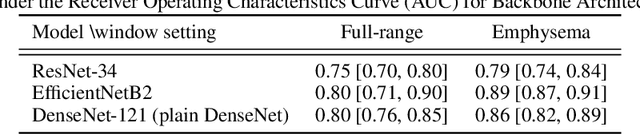
Abstract:Chronic Obstructive Pulmonary Disease (COPD) is a leading cause of death worldwide, yet early detection and treatment can prevent the progression of the disease. In contrast to the conventional method of detecting COPD with spirometry tests, X-ray Computed Tomography (CT) scans of the chest provide a measure of morphological changes in the lung. It has been shown that automated detection of COPD can be performed with deep learning models. However, the potential of incorporating optimal window setting selection, typically carried out by clinicians during examination of CT scans for COPD, is generally overlooked in deep learning approaches. We aim to optimize the binary classification of COPD with densely connected convolutional neural networks (DenseNets) through implementation of manual and automated Window-Setting Optimization (WSO) steps. Our dataset consisted of 78 CT scans from the Klinikum rechts der Isar research hospital. Repeated inference on the test set showed that without WSO, the plain DenseNet resulted in a mean slice-level AUC of 0.80$\pm$0.05. With input images manually adjusted to the emphysema window setting, the plain DenseNet model predicted COPD with a mean AUC of 0.86$\pm$0.04. By automating the WSO through addition of a customized layer to the DenseNet, an optimal window setting in the proximity of the emphysema window setting was learned and a mean AUC of 0.82$\pm$0.04 was achieved. Detection of COPD with DenseNet models was optimized by WSO of CT data to the emphysema window setting range, demonstrating the importance of implementing optimal window setting selection in the deep learning pipeline.
Practical Phase Retrieval Using Double Deep Image Priors
Nov 02, 2022Abstract:Phase retrieval (PR) concerns the recovery of complex phases from complex magnitudes. We identify the connection between the difficulty level and the number and variety of symmetries in PR problems. We focus on the most difficult far-field PR (FFPR), and propose a novel method using double deep image priors. In realistic evaluation, our method outperforms all competing methods by large margins. As a single-instance method, our method requires no training data and minimal hyperparameter tuning, and hence enjoys good practicality.
Phase Retrieval using Single-Instance Deep Generative Prior
Jun 22, 2021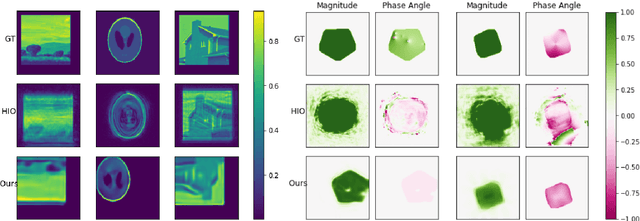
Abstract:Several deep learning methods for phase retrieval exist, but most of them fail on realistic data without precise support information. We propose a novel method based on single-instance deep generative prior that works well on complex-valued crystal data.
The Liver Tumor Segmentation Benchmark (LiTS)
Jan 13, 2019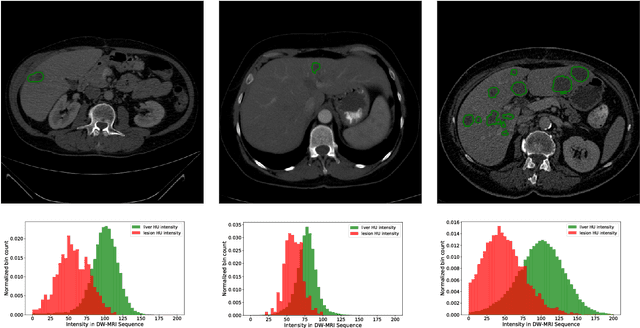
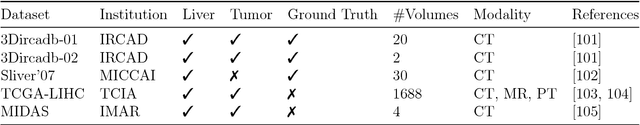
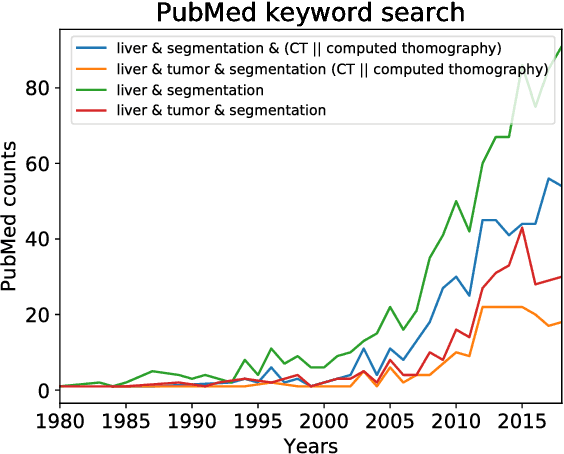
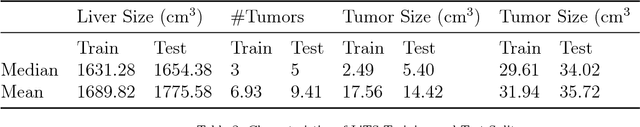
Abstract:In this work, we report the set-up and results of the Liver Tumor Segmentation Benchmark (LITS) organized in conjunction with the IEEE International Symposium on Biomedical Imaging (ISBI) 2016 and International Conference On Medical Image Computing Computer Assisted Intervention (MICCAI) 2017. Twenty four valid state-of-the-art liver and liver tumor segmentation algorithms were applied to a set of 131 computed tomography (CT) volumes with different types of tumor contrast levels (hyper-/hypo-intense), abnormalities in tissues (metastasectomie) size and varying amount of lesions. The submitted algorithms have been tested on 70 undisclosed volumes. The dataset is created in collaboration with seven hospitals and research institutions and manually reviewed by independent three radiologists. We found that not a single algorithm performed best for liver and tumors. The best liver segmentation algorithm achieved a Dice score of 0.96(MICCAI) whereas for tumor segmentation the best algorithm evaluated at 0.67(ISBI) and 0.70(MICCAI). The LITS image data and manual annotations continue to be publicly available through an online evaluation system as an ongoing benchmarking resource.
Automatic Liver and Tumor Segmentation of CT and MRI Volumes using Cascaded Fully Convolutional Neural Networks
Feb 23, 2017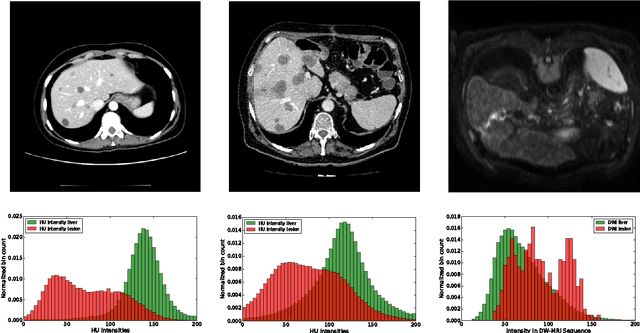
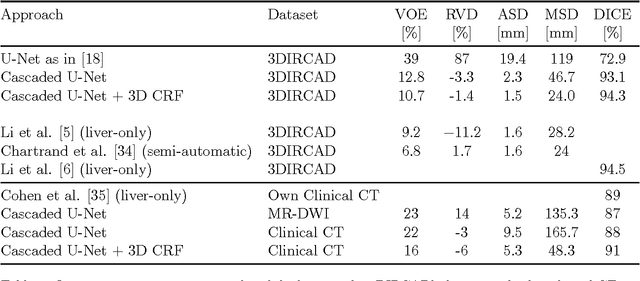
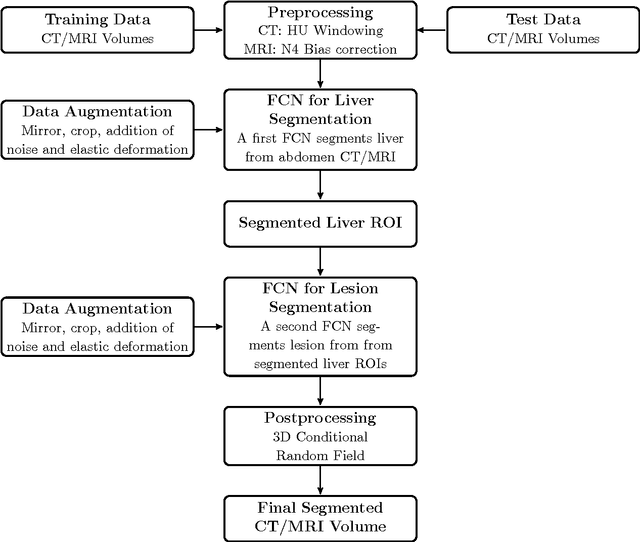
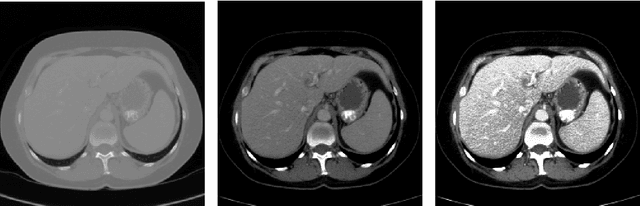
Abstract:Automatic segmentation of the liver and hepatic lesions is an important step towards deriving quantitative biomarkers for accurate clinical diagnosis and computer-aided decision support systems. This paper presents a method to automatically segment liver and lesions in CT and MRI abdomen images using cascaded fully convolutional neural networks (CFCNs) enabling the segmentation of a large-scale medical trial or quantitative image analysis. We train and cascade two FCNs for a combined segmentation of the liver and its lesions. In the first step, we train a FCN to segment the liver as ROI input for a second FCN. The second FCN solely segments lesions within the predicted liver ROIs of step 1. CFCN models were trained on an abdominal CT dataset comprising 100 hepatic tumor volumes. Validations on further datasets show that CFCN-based semantic liver and lesion segmentation achieves Dice scores over 94% for liver with computation times below 100s per volume. We further experimentally demonstrate the robustness of the proposed method on an 38 MRI liver tumor volumes and the public 3DIRCAD dataset.
Automatic Liver and Lesion Segmentation in CT Using Cascaded Fully Convolutional Neural Networks and 3D Conditional Random Fields
Oct 07, 2016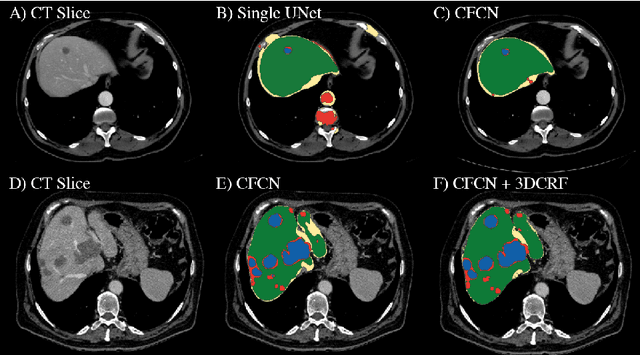
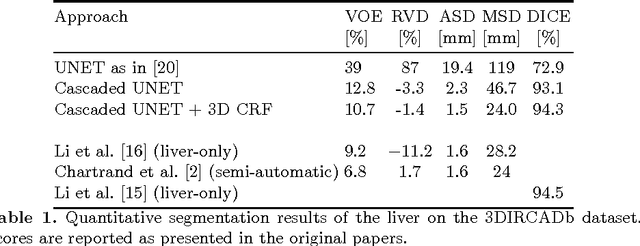
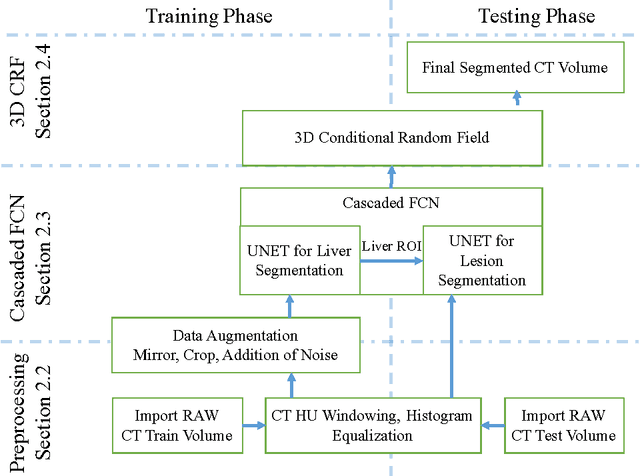
Abstract:Automatic segmentation of the liver and its lesion is an important step towards deriving quantitative biomarkers for accurate clinical diagnosis and computer-aided decision support systems. This paper presents a method to automatically segment liver and lesions in CT abdomen images using cascaded fully convolutional neural networks (CFCNs) and dense 3D conditional random fields (CRFs). We train and cascade two FCNs for a combined segmentation of the liver and its lesions. In the first step, we train a FCN to segment the liver as ROI input for a second FCN. The second FCN solely segments lesions from the predicted liver ROIs of step 1. We refine the segmentations of the CFCN using a dense 3D CRF that accounts for both spatial coherence and appearance. CFCN models were trained in a 2-fold cross-validation on the abdominal CT dataset 3DIRCAD comprising 15 hepatic tumor volumes. Our results show that CFCN-based semantic liver and lesion segmentation achieves Dice scores over 94% for liver with computation times below 100s per volume. We experimentally demonstrate the robustness of the proposed method as a decision support system with a high accuracy and speed for usage in daily clinical routine.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge