Chongyu Wang
GUI-Rise: Structured Reasoning and History Summarization for GUI Navigation
Oct 31, 2025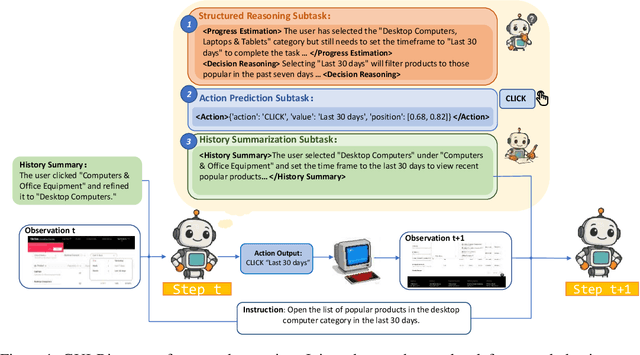
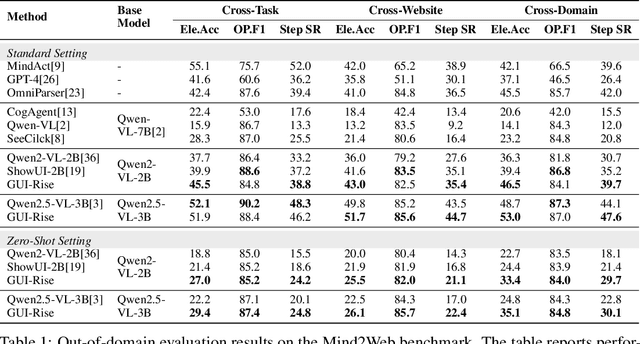
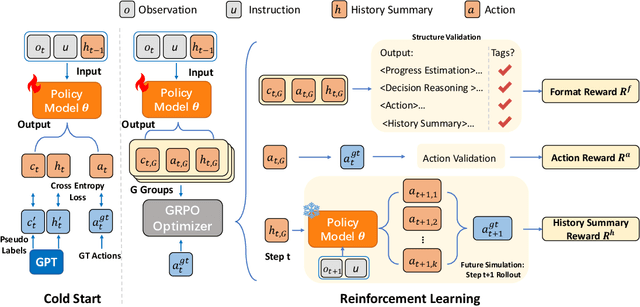
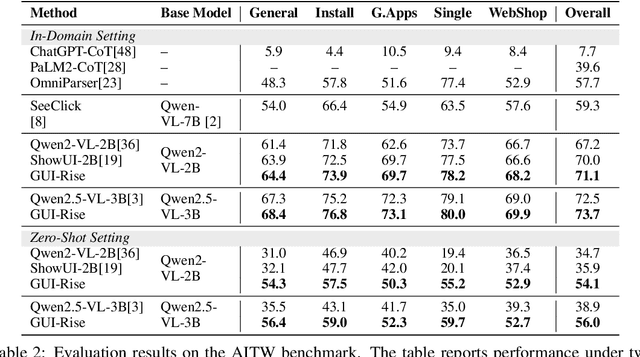
Abstract:While Multimodal Large Language Models (MLLMs) have advanced GUI navigation agents, current approaches face limitations in cross-domain generalization and effective history utilization. We present a reasoning-enhanced framework that systematically integrates structured reasoning, action prediction, and history summarization. The structured reasoning component generates coherent Chain-of-Thought analyses combining progress estimation and decision reasoning, which inform both immediate action predictions and compact history summaries for future steps. Based on this framework, we train a GUI agent, \textbf{GUI-Rise}, through supervised fine-tuning on pseudo-labeled trajectories and reinforcement learning with Group Relative Policy Optimization (GRPO). This framework employs specialized rewards, including a history-aware objective, directly linking summary quality to subsequent action performance. Comprehensive evaluations on standard benchmarks demonstrate state-of-the-art results under identical training data conditions, with particularly strong performance in out-of-domain scenarios. These findings validate our framework's ability to maintain robust reasoning and generalization across diverse GUI navigation tasks. Code is available at https://leon022.github.io/GUI-Rise.
Incremental Human-Object Interaction Detection with Invariant Relation Representation Learning
Oct 30, 2025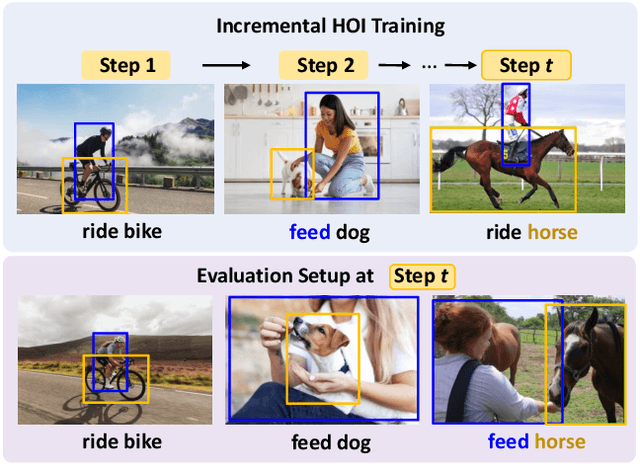
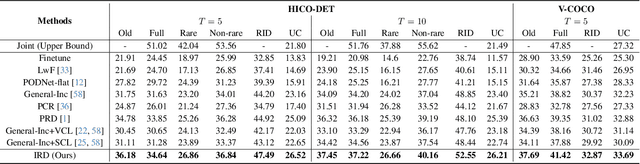
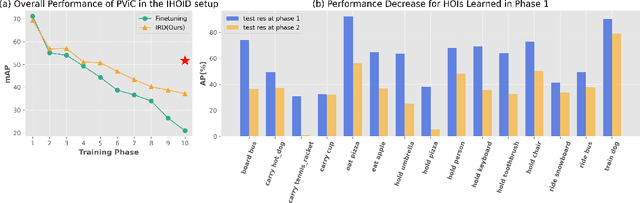
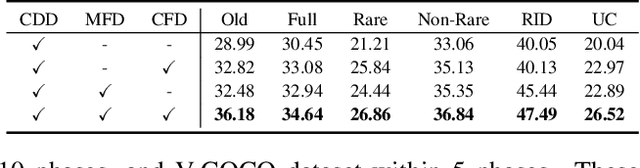
Abstract:In open-world environments, human-object interactions (HOIs) evolve continuously, challenging conventional closed-world HOI detection models. Inspired by humans' ability to progressively acquire knowledge, we explore incremental HOI detection (IHOID) to develop agents capable of discerning human-object relations in such dynamic environments. This setup confronts not only the common issue of catastrophic forgetting in incremental learning but also distinct challenges posed by interaction drift and detecting zero-shot HOI combinations with sequentially arriving data. Therefore, we propose a novel exemplar-free incremental relation distillation (IRD) framework. IRD decouples the learning of objects and relations, and introduces two unique distillation losses for learning invariant relation features across different HOI combinations that share the same relation. Extensive experiments on HICO-DET and V-COCO datasets demonstrate the superiority of our method over state-of-the-art baselines in mitigating forgetting, strengthening robustness against interaction drift, and generalization on zero-shot HOIs. Code is available at \href{https://github.com/weiyana/ContinualHOI}{this HTTP URL}
Advancing the Understanding of Fine-Grained 3D Forest Structures using Digital Cousins and Simulation-to-Reality: Methods and Datasets
Jan 07, 2025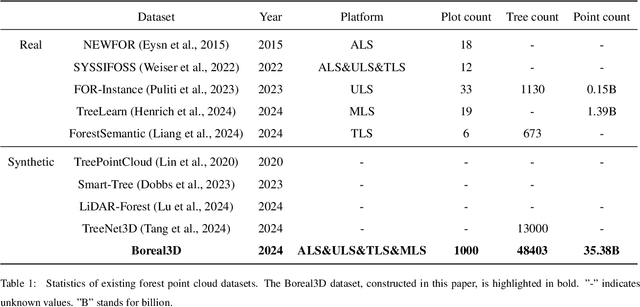
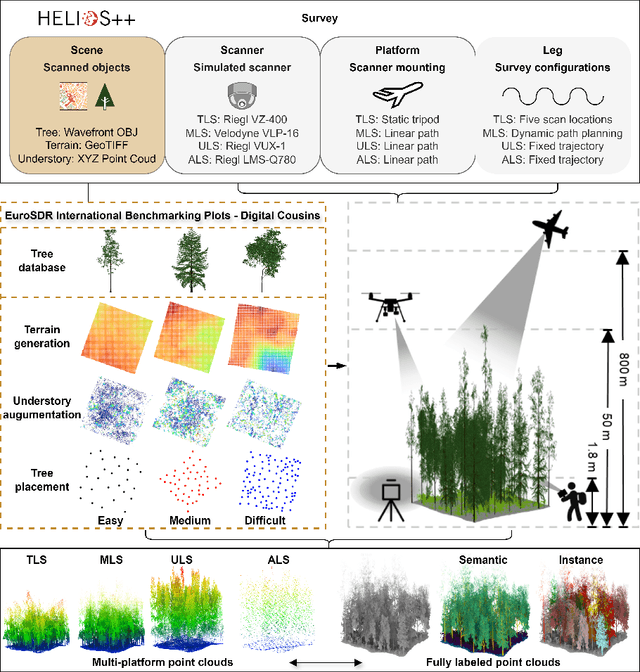
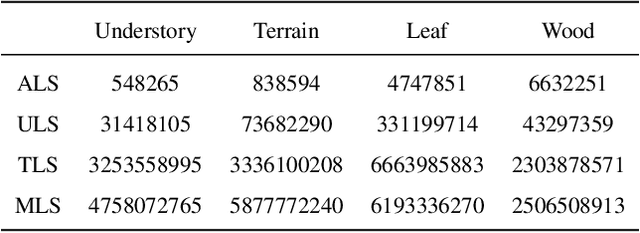
Abstract:Understanding and analyzing the spatial semantics and structure of forests is essential for accurate forest resource monitoring and ecosystem research. However, the lack of large-scale and annotated datasets has limited the widespread use of advanced intelligent techniques in this field. To address this challenge, a fully automated synthetic data generation and processing framework based on the concepts of Digital Cousins and Simulation-to-Reality (Sim2Real) is proposed, offering versatility and scalability to any size and platform. Using this process, we created the Boreal3D, the world's largest forest point cloud dataset. It includes 1000 highly realistic and structurally diverse forest plots across four different platforms, totaling 48,403 trees and over 35.3 billion points. Each point is labeled with semantic, instance, and viewpoint information, while each tree is described with structural parameters such as diameter, crown width, leaf area, and total volume. We designed and conducted extensive experiments to evaluate the potential of Boreal3D in advancing fine-grained 3D forest structure analysis in real-world applications. The results demonstrate that with certain strategies, models pre-trained on synthetic data can significantly improve performance when applied to real forest datasets. Especially, the findings reveal that fine-tuning with only 20% of real-world data enables the model to achieve performance comparable to models trained exclusively on entire real-world data, highlighting the value and potential of our proposed framework. The Boreal3D dataset, and more broadly, the synthetic data augmentation framework, is poised to become a critical resource for advancing research in large-scale 3D forest scene understanding and structural parameter estimation.
Relation-aware Hierarchical Prompt for Open-vocabulary Scene Graph Generation
Dec 26, 2024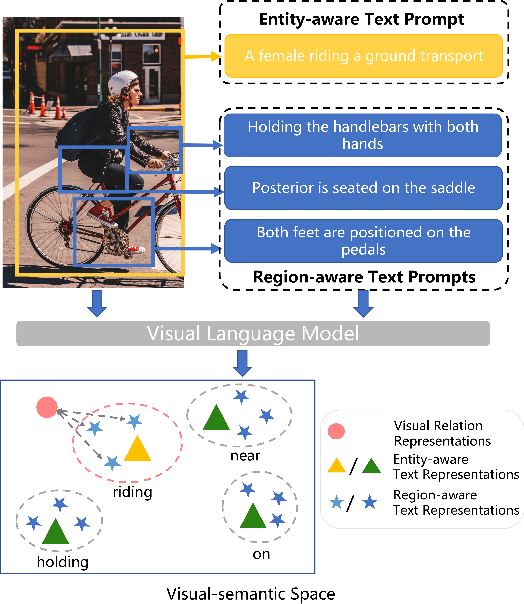
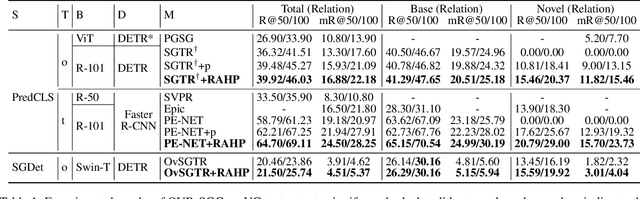
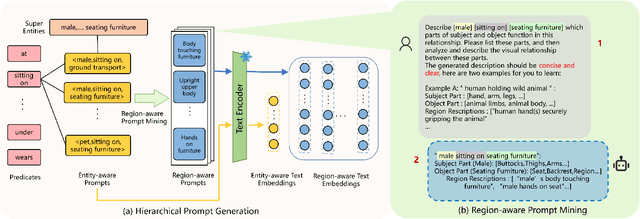
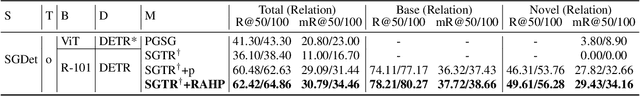
Abstract:Open-vocabulary Scene Graph Generation (OV-SGG) overcomes the limitations of the closed-set assumption by aligning visual relationship representations with open-vocabulary textual representations. This enables the identification of novel visual relationships, making it applicable to real-world scenarios with diverse relationships. However, existing OV-SGG methods are constrained by fixed text representations, limiting diversity and accuracy in image-text alignment. To address these challenges, we propose the Relation-Aware Hierarchical Prompting (RAHP) framework, which enhances text representation by integrating subject-object and region-specific relation information. Our approach utilizes entity clustering to address the complexity of relation triplet categories, enabling the effective integration of subject-object information. Additionally, we utilize a large language model (LLM) to generate detailed region-aware prompts, capturing fine-grained visual interactions and improving alignment between visual and textual modalities. RAHP also introduces a dynamic selection mechanism within Vision-Language Models (VLMs), which adaptively selects relevant text prompts based on the visual content, reducing noise from irrelevant prompts. Extensive experiments on the Visual Genome and Open Images v6 datasets demonstrate that our framework consistently achieves state-of-the-art performance, demonstrating its effectiveness in addressing the challenges of open-vocabulary scene graph generation.
Efficiently Expanding Receptive Fields: Local Split Attention and Parallel Aggregation for Enhanced Large-scale Point Cloud Semantic Segmentation
Sep 03, 2024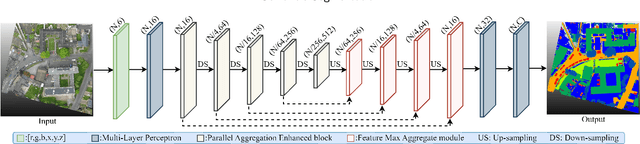
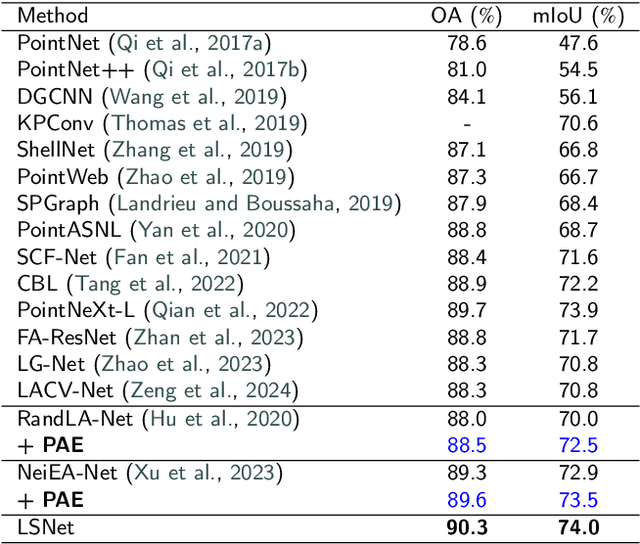
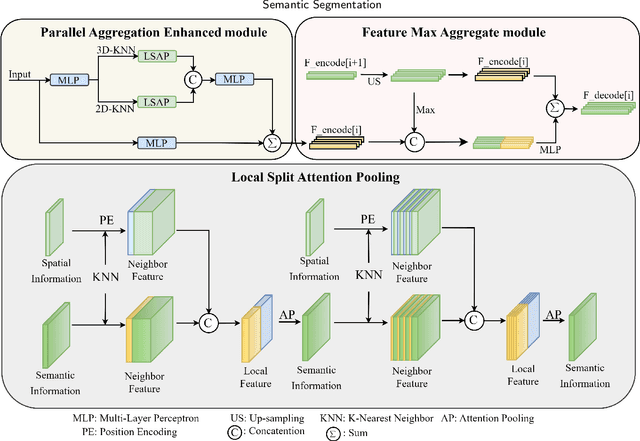
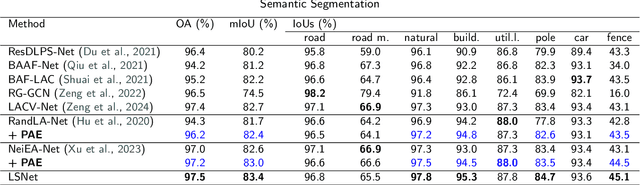
Abstract:Expanding the receptive field in a deep learning model for large-scale 3D point cloud segmentation is an effective technique for capturing rich contextual information, which consequently enhances the network's ability to learn meaningful features. However, this often leads to increased computational complexity and risk of overfitting, challenging the efficiency and effectiveness of the learning paradigm. To address these limitations, we propose the Local Split Attention Pooling (LSAP) mechanism to effectively expand the receptive field through a series of local split operations, thus facilitating the acquisition of broader contextual knowledge. Concurrently, it optimizes the computational workload associated with attention-pooling layers to ensure a more streamlined processing workflow. Based on LSAP, a Parallel Aggregation Enhancement (PAE) module is introduced to enable parallel processing of data using both 2D and 3D neighboring information to further enhance contextual representations within the network. In light of the aforementioned designs, we put forth a novel framework, designated as LSNet, for large-scale point cloud semantic segmentation. Extensive evaluations demonstrated the efficacy of seamlessly integrating the proposed PAE module into existing frameworks, yielding significant improvements in mean intersection over union (mIoU) metrics, with a notable increase of up to 11%. Furthermore, LSNet demonstrated superior performance compared to state-of-the-art semantic segmentation networks on three benchmark datasets, including S3DIS, Toronto3D, and SensatUrban. It is noteworthy that our method achieved a substantial speedup of approximately 38.8% compared to those employing similar-sized receptive fields, which serves to highlight both its computational efficiency and practical utility in real-world large-scale scenes.
Scalable Face Image Coding via StyleGAN Prior: Towards Compression for Human-Machine Collaborative Vision
Dec 25, 2023



Abstract:The accelerated proliferation of visual content and the rapid development of machine vision technologies bring significant challenges in delivering visual data on a gigantic scale, which shall be effectively represented to satisfy both human and machine requirements. In this work, we investigate how hierarchical representations derived from the advanced generative prior facilitate constructing an efficient scalable coding paradigm for human-machine collaborative vision. Our key insight is that by exploiting the StyleGAN prior, we can learn three-layered representations encoding hierarchical semantics, which are elaborately designed into the basic, middle, and enhanced layers, supporting machine intelligence and human visual perception in a progressive fashion. With the aim of achieving efficient compression, we propose the layer-wise scalable entropy transformer to reduce the redundancy between layers. Based on the multi-task scalable rate-distortion objective, the proposed scheme is jointly optimized to achieve optimal machine analysis performance, human perception experience, and compression ratio. We validate the proposed paradigm's feasibility in face image compression. Extensive qualitative and quantitative experimental results demonstrate the superiority of the proposed paradigm over the latest compression standard Versatile Video Coding (VVC) in terms of both machine analysis as well as human perception at extremely low bitrates ($<0.01$ bpp), offering new insights for human-machine collaborative compression.
Knowledge Graph Completion based on Tensor Decomposition for Disease Gene Prediction
Feb 18, 2023



Abstract:Accurate identification of disease genes has consistently been one of the keys to decoding a disease's molecular mechanism. Most current approaches focus on constructing biological networks and utilizing machine learning, especially, deep learning to identify disease genes, but ignore the complex relations between entities in the biological knowledge graph. In this paper, we construct a biological knowledge graph centered on diseases and genes, and develop an end-to-end Knowledge graph completion model for Disease Gene Prediction using interactional tensor decomposition (called KDGene). KDGene introduces an interaction module between the embeddings of entities and relations to tensor decomposition, which can effectively enhance the information interaction in biological knowledge. Experimental results show that KDGene significantly outperforms state-of-the-art algorithms. Furthermore, the comprehensive biological analysis of the case of diabetes mellitus confirms KDGene's ability for identifying new and accurate candidate genes. This work proposes a scalable knowledge graph completion framework to identify disease candidate genes, from which the results are promising to provide valuable references for further wet experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge