Bulat Ibragimov
rNCA: Self-Repairing Segmentation Masks
Dec 15, 2025Abstract:Accurately predicting topologically correct masks remains a difficult task for general segmentation models, which often produce fragmented or disconnected outputs. Fixing these artifacts typically requires hand-crafted refinement rules or architectures specialized to a particular task. Here, we show that Neural Cellular Automata (NCA) can be directly re-purposed as an effective refinement mechanism, using local, iterative updates guided by image context to repair segmentation masks. By training on imperfect masks and ground truths, the automaton learns the structural properties of the target shape while relying solely on local information. When applied to coarse, globally predicted masks, the learned dynamics progressively reconnect broken regions, prune loose fragments and converge towards stable, topologically consistent results. We show how refinement NCA (rNCA) can be easily applied to repair common topological errors produced by different base segmentation models and tasks: for fragmented retinal vessels, it yields 2-3% gains in Dice/clDice and improves Betti errors, reducing $β_0$ errors by 60% and $β_1$ by 20%; for myocardium, it repairs 61.5% of broken cases in a zero-shot setting while lowering ASSD and HD by 19% and 16%, respectively. This showcases NCA as effective and broadly applicable refiners.
CARMA: Collocation-Aware Resource Manager with GPU Memory Estimator
Aug 26, 2025Abstract:Studies conducted on enterprise-scale infrastructure have shown that GPUs -- the core computational resource for deep learning (DL) training -- are often significantly underutilized. DL task collocation on GPUs is an opportunity to address this challenge. However, it may result in (1) out-of-memory crashes for the subsequently arriving task and (2) slowdowns for all tasks sharing the GPU due to resource interference. The former challenge poses a threat to robustness, while the latter affects the quality of service and energy efficiency. We propose CARMA, a server-scale task-level collocation-aware resource management system that handles both collocation challenges. CARMA encompasses GPUMemNet, a novel ML-based GPU memory estimator framework for DL training tasks, to minimize out-of-memory errors and introduces collocation policies that cap GPU utilization to minimize interference. Furthermore, CARMA introduces a recovery method to ensure robust restart of tasks that crash. Our evaluation on traces modeled after real-world DL training task traces shows that CARMA increases the GPU utilization over time by 39.3\%, decreases the end-to-end execution time by $\sim$26.7\%, and reduces the GPU energy use by $\sim$14.2\%.
Gaze-Assisted Medical Image Segmentation
Oct 23, 2024Abstract:The annotation of patient organs is a crucial part of various diagnostic and treatment procedures, such as radiotherapy planning. Manual annotation is extremely time-consuming, while its automation using modern image analysis techniques has not yet reached levels sufficient for clinical adoption. This paper investigates the idea of semi-supervised medical image segmentation using human gaze as interactive input for segmentation correction. In particular, we fine-tuned the Segment Anything Model in Medical Images (MedSAM), a public solution that uses various prompt types as additional input for semi-automated segmentation correction. We used human gaze data from reading abdominal images as a prompt for fine-tuning MedSAM. The model was validated on a public WORD database, which consists of 120 CT scans of 16 abdominal organs. The results of the gaze-assisted MedSAM were shown to be superior to the results of the state-of-the-art segmentation models. In particular, the average Dice coefficient for 16 abdominal organs was 85.8%, 86.7%, 81.7%, and 90.5% for nnUNetV2, ResUNet, original MedSAM, and our gaze-assisted MedSAM model, respectively.
Building an AI Support Tool for Real-time Ulcerative Colitis Diagnosis
Apr 10, 2024Abstract:Ulcerative Colitis (UC) is a chronic inflammatory bowel disease decreasing life quality through symptoms such as bloody diarrhoea and abdominal pain. Endoscopy is a cornerstone of diagnosis and monitoring of UC. The Mayo endoscopic subscore (MES) index is the standard for measuring UC severity during endoscopic evaluation. However, the MES is subject to high inter-observer variability leading to misdiagnosis and suboptimal treatment. We propose using a machine-learning based MES classification system to support the endoscopic process and to mitigate the observer-variability. The system runs real-time in the clinic and augments doctors' decision-making during the endoscopy. This project report outlines the process of designing, creating and evaluating our system. We describe our initial evaluation, which is a combination of a standard non-clinical model test and a first clinical test of the system on a real patient.
Cross-Modal Conceptualization in Bottleneck Models
Oct 23, 2023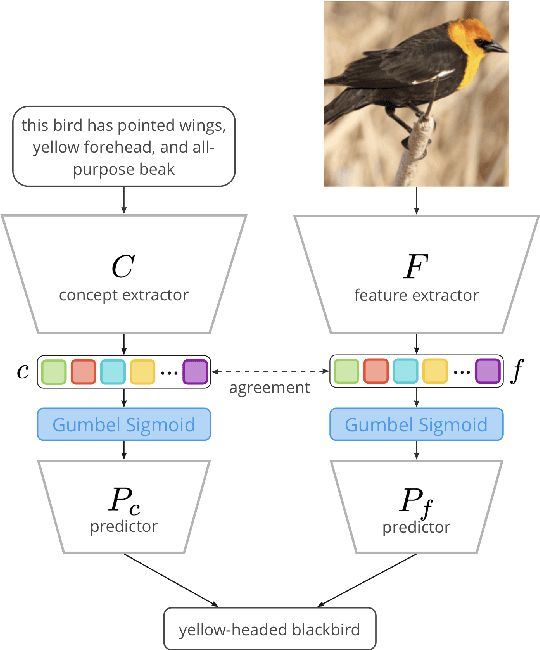
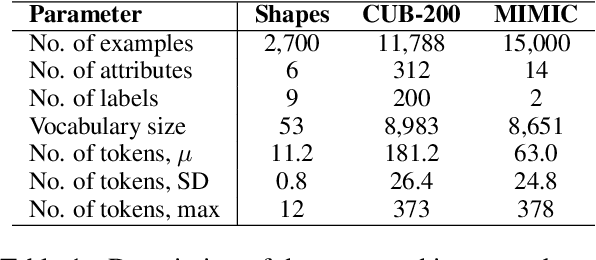
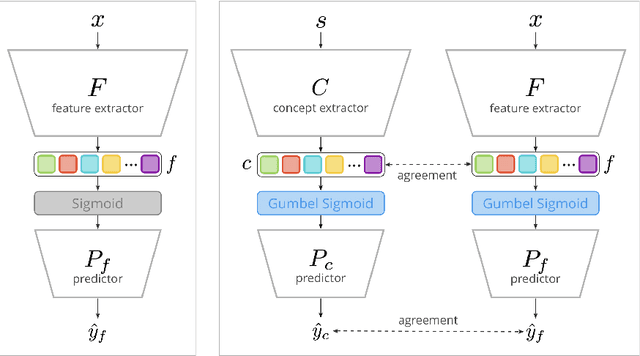
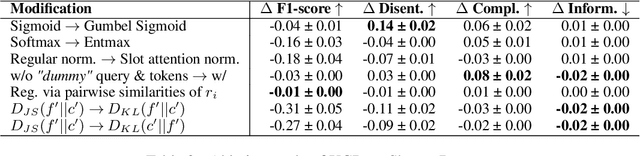
Abstract:Concept Bottleneck Models (CBMs) assume that training examples (e.g., x-ray images) are annotated with high-level concepts (e.g., types of abnormalities), and perform classification by first predicting the concepts, followed by predicting the label relying on these concepts. The main difficulty in using CBMs comes from having to choose concepts that are predictive of the label and then having to label training examples with these concepts. In our approach, we adopt a more moderate assumption and instead use text descriptions (e.g., radiology reports), accompanying the images in training, to guide the induction of concepts. Our cross-modal approach treats concepts as discrete latent variables and promotes concepts that (1) are predictive of the label, and (2) can be predicted reliably from both the image and text. Through experiments conducted on datasets ranging from synthetic datasets (e.g., synthetic images with generated descriptions) to realistic medical imaging datasets, we demonstrate that cross-modal learning encourages the induction of interpretable concepts while also facilitating disentanglement. Our results also suggest that this guidance leads to increased robustness by suppressing the reliance on shortcut features.
A Sequential Framework for Detection and Classification of Abnormal Teeth in Panoramic X-rays
Sep 04, 2023Abstract:This paper describes our solution for the Dental Enumeration and Diagnosis on Panoramic X-rays Challenge at MICCAI 2023. Our approach consists of a multi-step framework tailored to the task of detecting and classifying abnormal teeth. The solution includes three sequential stages: dental instance detection, healthy instance filtering, and abnormal instance classification. In the first stage, we employed a Faster-RCNN model for detecting and identifying teeth. In subsequent stages, we designed a model that merged the encoding pathway of a pretrained U-net, optimized for dental lesion detection, with the Vgg16 architecture. The resulting model was first used for filtering out healthy teeth. Then, any identified abnormal teeth were categorized, potentially falling into one or more of the following conditions: embedded, periapical lesion, caries, deep caries. The model performing dental instance detection achieved an AP score of 0.49. The model responsible for identifying healthy teeth attained an F1 score of 0.71. Meanwhile, the model trained for multi-label dental disease classification achieved an F1 score of 0.76. The code is available at https://github.com/tudordascalu/2d-teeth-detection-challenge.
3DTeethSeg'22: 3D Teeth Scan Segmentation and Labeling Challenge
May 29, 2023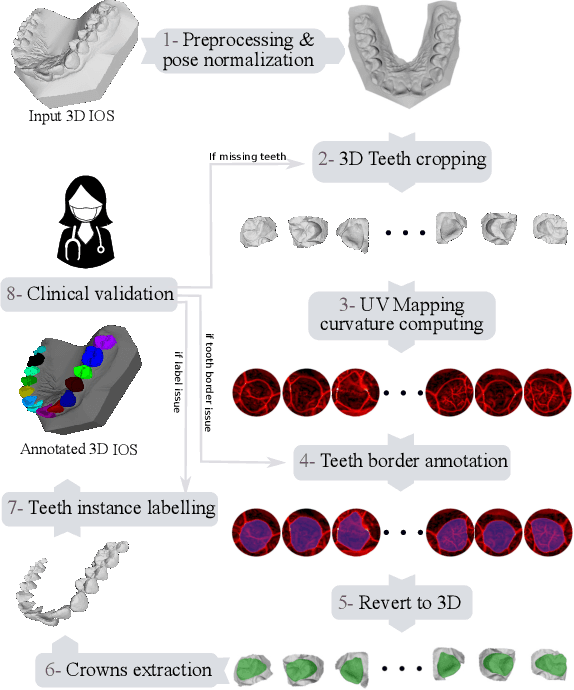
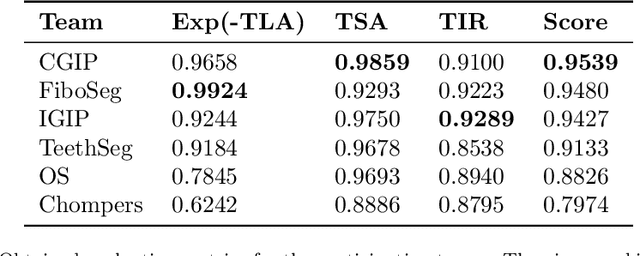
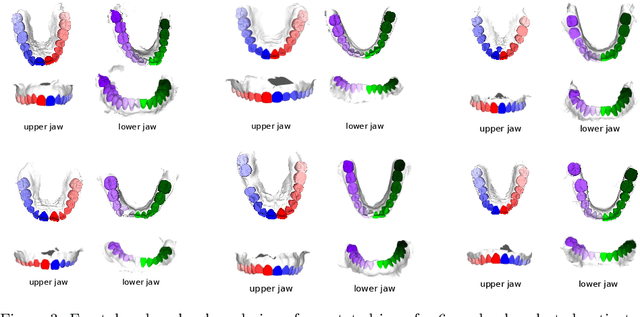
Abstract:Teeth localization, segmentation, and labeling from intra-oral 3D scans are essential tasks in modern dentistry to enhance dental diagnostics, treatment planning, and population-based studies on oral health. However, developing automated algorithms for teeth analysis presents significant challenges due to variations in dental anatomy, imaging protocols, and limited availability of publicly accessible data. To address these challenges, the 3DTeethSeg'22 challenge was organized in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) in 2022, with a call for algorithms tackling teeth localization, segmentation, and labeling from intraoral 3D scans. A dataset comprising a total of 1800 scans from 900 patients was prepared, and each tooth was individually annotated by a human-machine hybrid algorithm. A total of 6 algorithms were evaluated on this dataset. In this study, we present the evaluation results of the 3DTeethSeg'22 challenge. The 3DTeethSeg'22 challenge code can be accessed at: https://github.com/abenhamadou/3DTeethSeg22_challenge
Improved automated lesion segmentation in whole-body FDG/PET-CT via Test-Time Augmentation
Oct 14, 2022
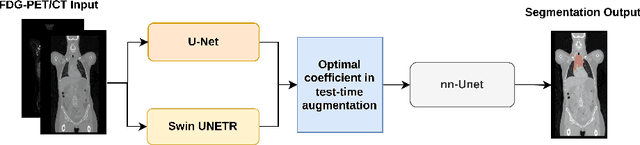
Abstract:Numerous oncology indications have extensively quantified metabolically active tumors using positron emission tomography (PET) and computed tomography (CT). F-fluorodeoxyglucose-positron emission tomography (FDG-PET) is frequently utilized in clinical practice and clinical drug research to detect and measure metabolically active malignancies. The assessment of tumor burden using manual or computer-assisted tumor segmentation in FDG-PET images is widespread. Deep learning algorithms have also produced effective solutions in this area. However, there may be a need to improve the performance of a pre-trained deep learning network without the opportunity to modify this network. We investigate the potential benefits of test-time augmentation for segmenting tumors from PET-CT pairings. We applied a new framework of multilevel and multimodal tumor segmentation techniques that can simultaneously consider PET and CT data. In this study, we improve the network using a learnable composition of test time augmentations. We trained U-Net and Swin U-Netr on the training database to determine how different test time augmentation improved segmentation performance. We also developed an algorithm that finds an optimal test time augmentation contribution coefficient set. Using the newly trained U-Net and Swin U-Netr results, we defined an optimal set of coefficients for test-time augmentation and utilized them in combination with a pre-trained fixed nnU-Net. The ultimate idea is to improve performance at the time of testing when the model is fixed. Averaging the predictions with varying ratios on the augmented data can improve prediction accuracy. Our code will be available at \url{https://github.com/sepidehamiri/pet\_seg\_unet}
NeuralSympCheck: A Symptom Checking and Disease Diagnostic Neural Model with Logic Regularization
Jun 02, 2022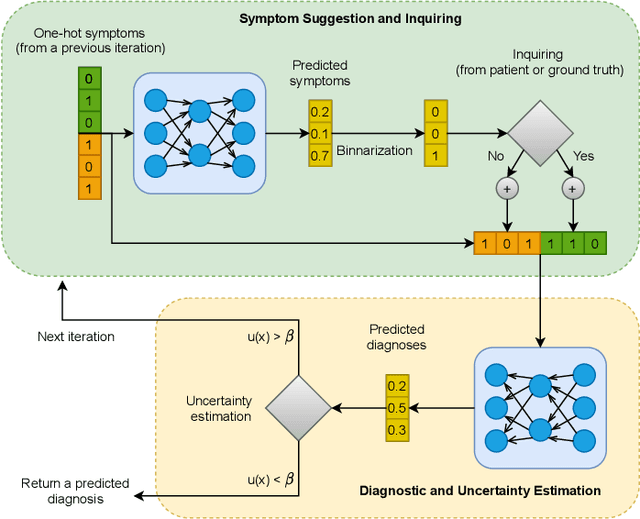
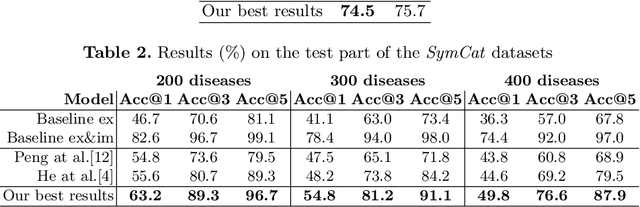
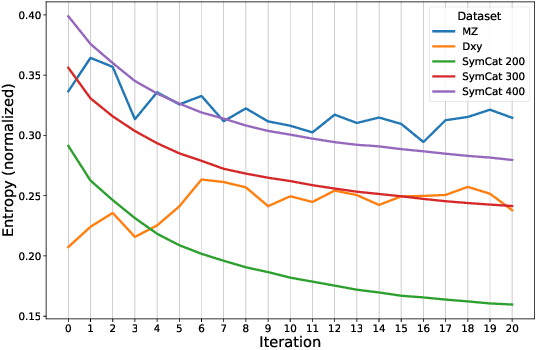
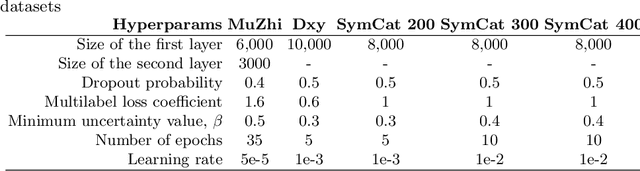
Abstract:The symptom checking systems inquire users for their symptoms and perform a rapid and affordable medical assessment of their condition. The basic symptom checking systems based on Bayesian methods, decision trees, or information gain methods are easy to train and do not require significant computational resources. However, their drawbacks are low relevance of proposed symptoms and insufficient quality of diagnostics. The best results on these tasks are achieved by reinforcement learning models. Their weaknesses are the difficulty of developing and training such systems and limited applicability to cases with large and sparse decision spaces. We propose a new approach based on the supervised learning of neural models with logic regularization that combines the advantages of the different methods. Our experiments on real and synthetic data show that the proposed approach outperforms the best existing methods in the accuracy of diagnosis when the number of diagnoses and symptoms is large.
Minimal Variance Sampling in Stochastic Gradient Boosting
Oct 29, 2019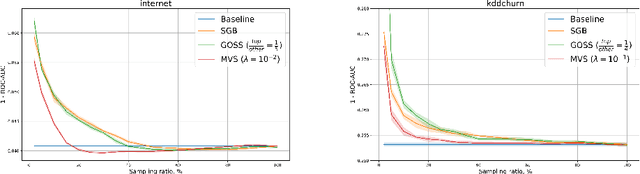
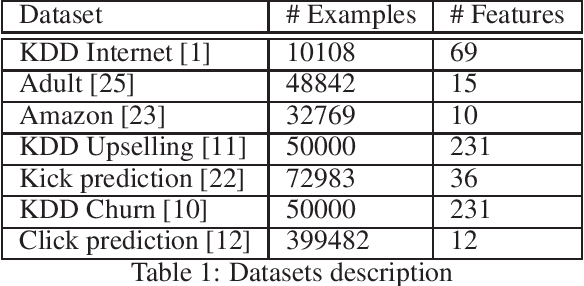
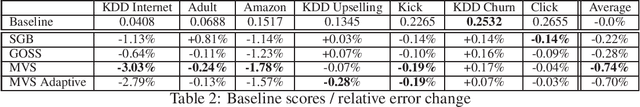
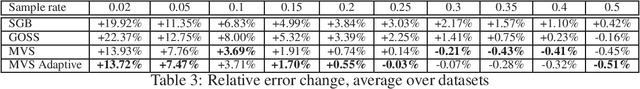
Abstract:Stochastic Gradient Boosting (SGB) is a widely used approach to regularization of boosting models based on decision trees. It was shown that, in many cases, random sampling at each iteration can lead to better generalization performance of the model and can also decrease the learning time. Different sampling approaches were proposed, where probabilities are not uniform, and it is not currently clear which approach is the most effective. In this paper, we formulate the problem of randomization in SGB in terms of optimization of sampling probabilities to maximize the estimation accuracy of split scoring used to train decision trees. This optimization problem has a closed-form nearly optimal solution, and it leads to a new sampling technique, which we call Minimal Variance Sampling (MVS). The method both decreases the number of examples needed for each iteration of boosting and increases the quality of the model significantly as compared to the state-of-the art sampling methods. The superiority of the algorithm was confirmed by introducing MVS as a new default option for subsampling in CatBoost, a gradient boosting library achieving state-of-the-art quality on various machine learning tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge