Sepideh Amiri
Improved automated lesion segmentation in whole-body FDG/PET-CT via Test-Time Augmentation
Oct 14, 2022
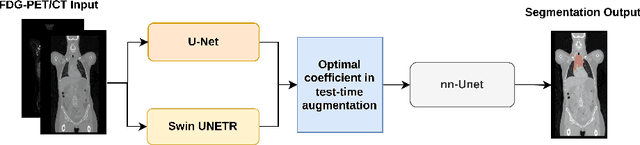
Abstract:Numerous oncology indications have extensively quantified metabolically active tumors using positron emission tomography (PET) and computed tomography (CT). F-fluorodeoxyglucose-positron emission tomography (FDG-PET) is frequently utilized in clinical practice and clinical drug research to detect and measure metabolically active malignancies. The assessment of tumor burden using manual or computer-assisted tumor segmentation in FDG-PET images is widespread. Deep learning algorithms have also produced effective solutions in this area. However, there may be a need to improve the performance of a pre-trained deep learning network without the opportunity to modify this network. We investigate the potential benefits of test-time augmentation for segmenting tumors from PET-CT pairings. We applied a new framework of multilevel and multimodal tumor segmentation techniques that can simultaneously consider PET and CT data. In this study, we improve the network using a learnable composition of test time augmentations. We trained U-Net and Swin U-Netr on the training database to determine how different test time augmentation improved segmentation performance. We also developed an algorithm that finds an optimal test time augmentation contribution coefficient set. Using the newly trained U-Net and Swin U-Netr results, we defined an optimal set of coefficients for test-time augmentation and utilized them in combination with a pre-trained fixed nnU-Net. The ultimate idea is to improve performance at the time of testing when the model is fixed. Averaging the predictions with varying ratios on the augmented data can improve prediction accuracy. Our code will be available at \url{https://github.com/sepidehamiri/pet\_seg\_unet}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge