Alexey Kornaev
Awareness of uncertainty in classification using a multivariate model and multi-views
Apr 16, 2024



Abstract:One of the ways to make artificial intelligence more natural is to give it some room for doubt. Two main questions should be resolved in that way. First, how to train a model to estimate uncertainties of its own predictions? And then, what to do with the uncertain predictions if they appear? First, we proposed an uncertainty-aware negative log-likelihood loss for the case of N-dimensional multivariate normal distribution with spherical variance matrix to the solution of N-classes classification tasks. The loss is similar to the heteroscedastic regression loss. The proposed model regularizes uncertain predictions, and trains to calculate both the predictions and their uncertainty estimations. The model fits well with the label smoothing technique. Second, we expanded the limits of data augmentation at the training and test stages, and made the trained model to give multiple predictions for a given number of augmented versions of each test sample. Given the multi-view predictions together with their uncertainties and confidences, we proposed several methods to calculate final predictions, including mode values and bin counts with soft and hard weights. For the latter method, we formalized the model tuning task in the form of multimodal optimization with non-differentiable criteria of maximum accuracy, and applied particle swarm optimization to solve the tuning task. The proposed methodology was tested using CIFAR-10 dataset with clean and noisy labels and demonstrated good results in comparison with other uncertainty estimation methods related to sample selection, co-teaching, and label smoothing.
Application of Deep Learning Methods to Processing of Noisy Medical Video Data
Apr 16, 2024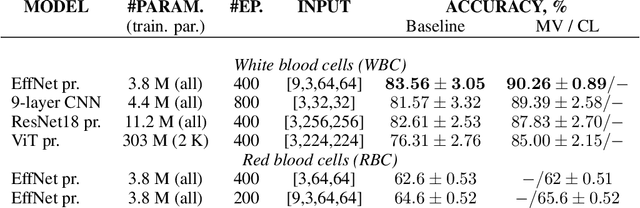



Abstract:Cells count become a challenging problem when the cells move in a continuous stream, and their boundaries are difficult for visual detection. To resolve this problem we modified the training and decision making processes using curriculum learning and multi-view predictions techniques, respectively.
Cross-Modal Conceptualization in Bottleneck Models
Oct 23, 2023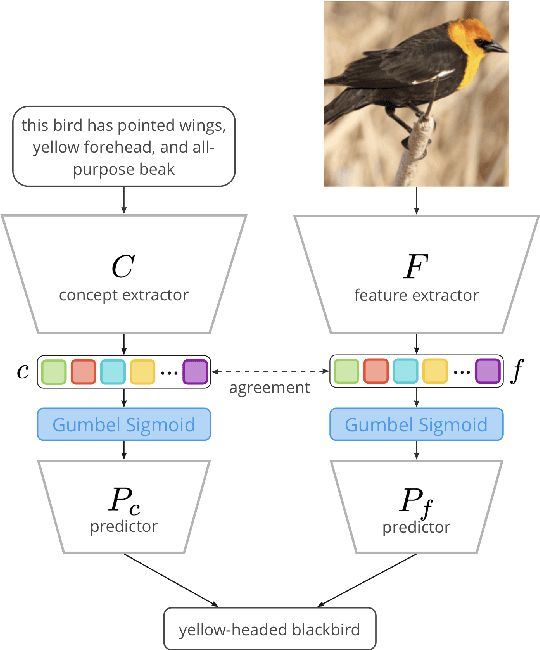
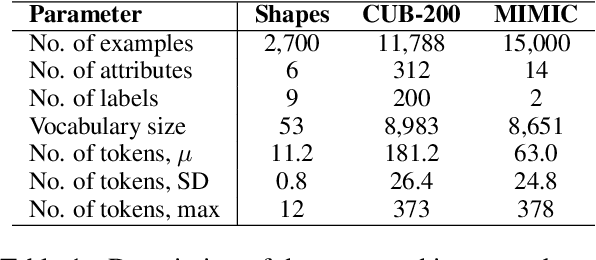
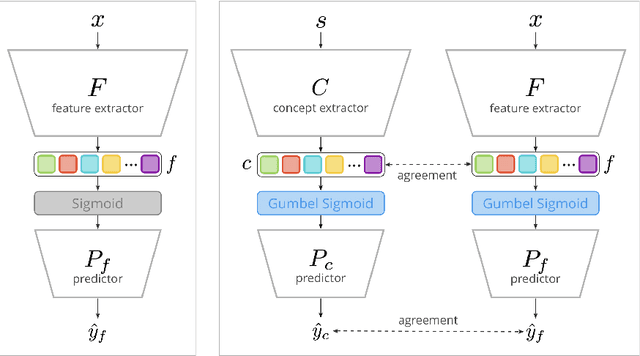
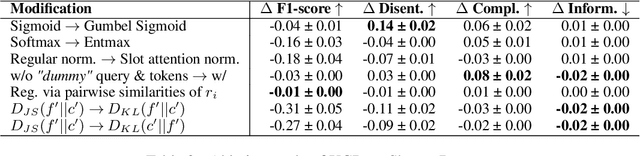
Abstract:Concept Bottleneck Models (CBMs) assume that training examples (e.g., x-ray images) are annotated with high-level concepts (e.g., types of abnormalities), and perform classification by first predicting the concepts, followed by predicting the label relying on these concepts. The main difficulty in using CBMs comes from having to choose concepts that are predictive of the label and then having to label training examples with these concepts. In our approach, we adopt a more moderate assumption and instead use text descriptions (e.g., radiology reports), accompanying the images in training, to guide the induction of concepts. Our cross-modal approach treats concepts as discrete latent variables and promotes concepts that (1) are predictive of the label, and (2) can be predicted reliably from both the image and text. Through experiments conducted on datasets ranging from synthetic datasets (e.g., synthetic images with generated descriptions) to realistic medical imaging datasets, we demonstrate that cross-modal learning encourages the induction of interpretable concepts while also facilitating disentanglement. Our results also suggest that this guidance leads to increased robustness by suppressing the reliance on shortcut features.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge