Aman Shrivastava
MedIL: Implicit Latent Spaces for Generating Heterogeneous Medical Images at Arbitrary Resolutions
Apr 12, 2025



Abstract:In this work, we introduce MedIL, a first-of-its-kind autoencoder built for encoding medical images with heterogeneous sizes and resolutions for image generation. Medical images are often large and heterogeneous, where fine details are of vital clinical importance. Image properties change drastically when considering acquisition equipment, patient demographics, and pathology, making realistic medical image generation challenging. Recent work in latent diffusion models (LDMs) has shown success in generating images resampled to a fixed-size. However, this is a narrow subset of the resolutions native to image acquisition, and resampling discards fine anatomical details. MedIL utilizes implicit neural representations to treat images as continuous signals, where encoding and decoding can be performed at arbitrary resolutions without prior resampling. We quantitatively and qualitatively show how MedIL compresses and preserves clinically-relevant features over large multi-site, multi-resolution datasets of both T1w brain MRIs and lung CTs. We further demonstrate how MedIL can influence the quality of images generated with a diffusion model, and discuss how MedIL can enhance generative models to resemble raw clinical acquisitions.
NASDM: Nuclei-Aware Semantic Histopathology Image Generation Using Diffusion Models
Mar 20, 2023



Abstract:In recent years, computational pathology has seen tremendous progress driven by deep learning methods in segmentation and classification tasks aiding prognostic and diagnostic settings. Nuclei segmentation, for instance, is an important task for diagnosing different cancers. However, training deep learning models for nuclei segmentation requires large amounts of annotated data, which is expensive to collect and label. This necessitates explorations into generative modeling of histopathological images. In this work, we use recent advances in conditional diffusion modeling to formulate a first-of-its-kind nuclei-aware semantic tissue generation framework (NASDM) which can synthesize realistic tissue samples given a semantic instance mask of up to six different nuclei types, enabling pixel-perfect nuclei localization in generated samples. These synthetic images are useful in applications in pathology pedagogy, validation of models, and supplementation of existing nuclei segmentation datasets. We demonstrate that NASDM is able to synthesize high-quality histopathology images of the colon with superior quality and semantic controllability over existing generative methods.
Improving Interpretability via Explicit Word Interaction Graph Layer
Feb 03, 2023



Abstract:Recent NLP literature has seen growing interest in improving model interpretability. Along this direction, we propose a trainable neural network layer that learns a global interaction graph between words and then selects more informative words using the learned word interactions. Our layer, we call WIGRAPH, can plug into any neural network-based NLP text classifiers right after its word embedding layer. Across multiple SOTA NLP models and various NLP datasets, we demonstrate that adding the WIGRAPH layer substantially improves NLP models' interpretability and enhances models' prediction performance at the same time.
* 15 pages, AAAI 2023
CLIP-Lite: Information Efficient Visual Representation Learning from Textual Annotations
Dec 14, 2021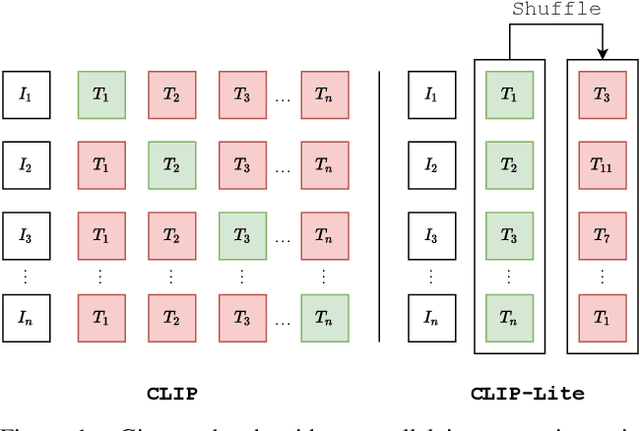
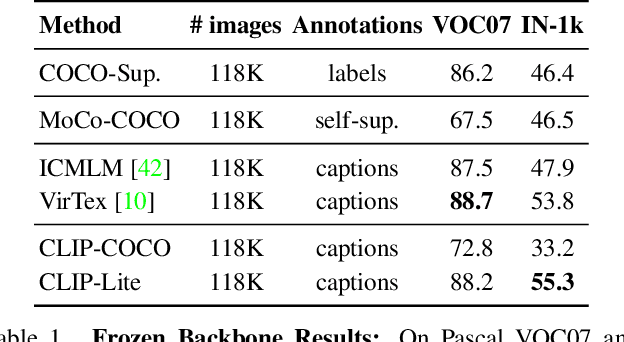

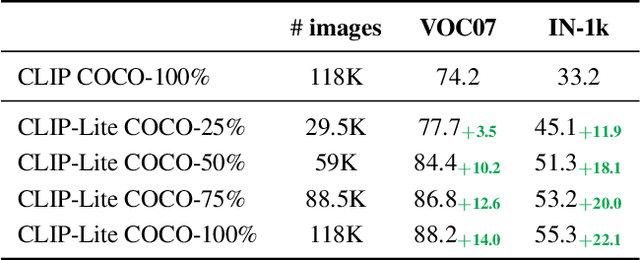
Abstract:We propose CLIP-Lite, an information efficient method for visual representation learning by feature alignment with textual annotations. Compared to the previously proposed CLIP model, CLIP-Lite requires only one negative image-text sample pair for every positive image-text sample during the optimization of its contrastive learning objective. We accomplish this by taking advantage of an information efficient lower-bound to maximize the mutual information between the two input modalities. This allows CLIP-Lite to be trained with significantly reduced amounts of data and batch sizes while obtaining better performance than CLIP. We evaluate CLIP-Lite by pretraining on the COCO-Captions dataset and testing transfer learning to other datasets. CLIP-Lite obtains a +15.4% mAP absolute gain in performance on Pascal VOC classification, and a +22.1% top-1 accuracy gain on ImageNet, while being comparable or superior to other, more complex, text-supervised models. CLIP-Lite is also superior to CLIP on image and text retrieval, zero-shot classification, and visual grounding. Finally, by performing explicit image-text alignment during representation learning, we show that CLIP-Lite can leverage language semantics to encourage bias-free visual representations that can be used in downstream tasks.
Estimating and Maximizing Mutual Information for Knowledge Distillation
Oct 29, 2021



Abstract:Knowledge distillation is a widely used general technique to transfer knowledge from a teacher network to a student network. In this work, we propose Mutual Information Maximization Knowledge Distillation (MIMKD). Our method uses a contrastive objective to simultaneously estimate and maximize a lower bound on the mutual information between intermediate and global feature representations from the teacher and the student networks. Our method is flexible, as the proposed mutual information maximization does not impose significant constraints on the structure of the intermediate features of the networks. As such, we can distill knowledge from arbitrary teachers to arbitrary students. Our empirical results show that our method outperforms competing approaches across a wide range of student-teacher pairs with different capacities, with different architectures, and when student networks are with extremely low capacity. We are able to obtain 74.55% accuracy on CIFAR100 with a ShufflenetV2 from a baseline accuracy of 69.8% by distilling knowledge from ResNet50.
Cluster-to-Conquer: A Framework for End-to-End Multi-Instance Learning for Whole Slide Image Classification
Mar 19, 2021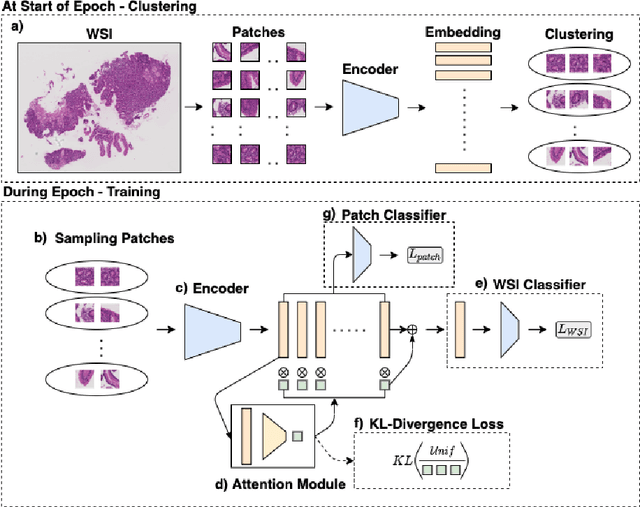
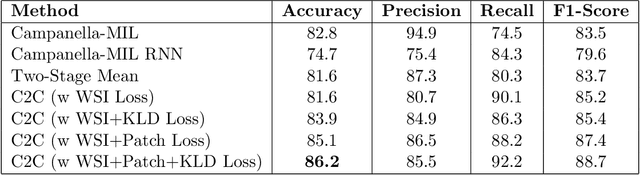
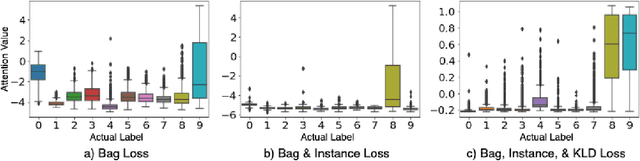
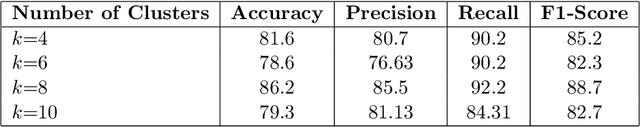
Abstract:In recent years, the availability of digitized Whole Slide Images (WSIs) has enabled the use of deep learning-based computer vision techniques for automated disease diagnosis. However, WSIs present unique computational and algorithmic challenges. WSIs are gigapixel-sized ($\sim$100K pixels), making them infeasible to be used directly for training deep neural networks. Also, often only slide-level labels are available for training as detailed annotations are tedious and can be time-consuming for experts. Approaches using multiple-instance learning (MIL) frameworks have been shown to overcome these challenges. Current state-of-the-art approaches divide the learning framework into two decoupled parts: a convolutional neural network (CNN) for encoding the patches followed by an independent aggregation approach for slide-level prediction. In this approach, the aggregation step has no bearing on the representations learned by the CNN encoder. We have proposed an end-to-end framework that clusters the patches from a WSI into ${k}$-groups, samples ${k}'$ patches from each group for training, and uses an adaptive attention mechanism for slide level prediction; Cluster-to-Conquer (C2C). We have demonstrated that dividing a WSI into clusters can improve the model training by exposing it to diverse discriminative features extracted from the patches. We regularized the clustering mechanism by introducing a KL-divergence loss between the attention weights of patches in a cluster and the uniform distribution. The framework is optimized end-to-end on slide-level cross-entropy, patch-level cross-entropy, and KL-divergence loss (Implementation: https://github.com/YashSharma/C2C).
Self-Attentive Adversarial Stain Normalization
Sep 04, 2019
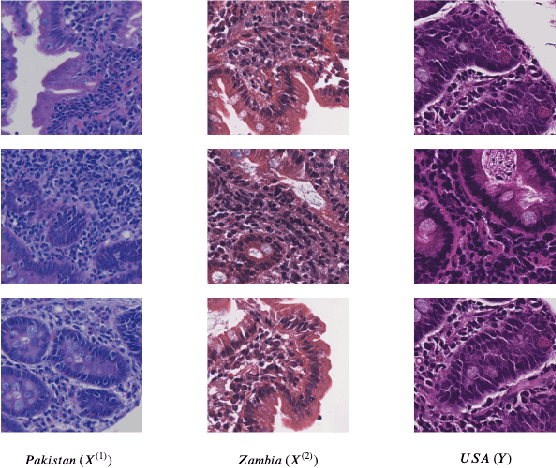
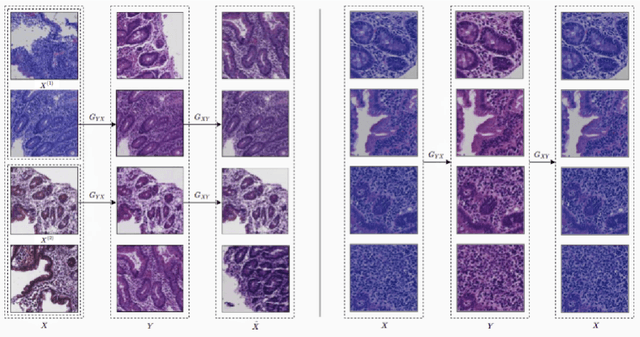

Abstract:Hematoxylin and Eosin (H&E) stained Whole Slide Images (WSIs) are utilized for biopsy visualization-based diagnostic and prognostic assessment of diseases. Variation in the H&E staining process across different lab sites can lead to significant variations in biopsy image appearance. These variations introduce an undesirable bias when the slides are examined by pathologists or used for training deep learning models. To reduce this bias, slides need to be translated to a common domain of stain appearance before analysis. We propose a Self-Attentive Adversarial Stain Normalization (SAASN) approach for the normalization of multiple stain appearances to a common domain. This unsupervised generative adversarial approach includes self-attention mechanism for synthesizing images with finer detail while preserving the structural consistency of the biopsy features during translation. SAASN demonstrates consistent and superior performance compared to other popular stain normalization techniques on H&E stained duodenal biopsy image data.
Deep Learning for Visual Recognition of Environmental Enteropathy and Celiac Disease
Aug 08, 2019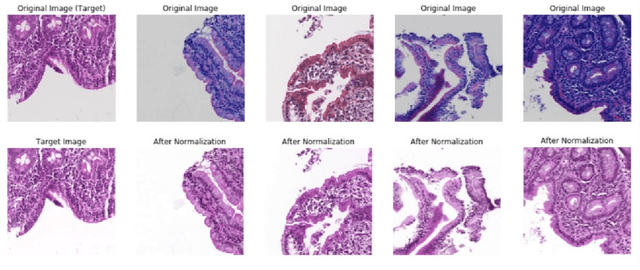
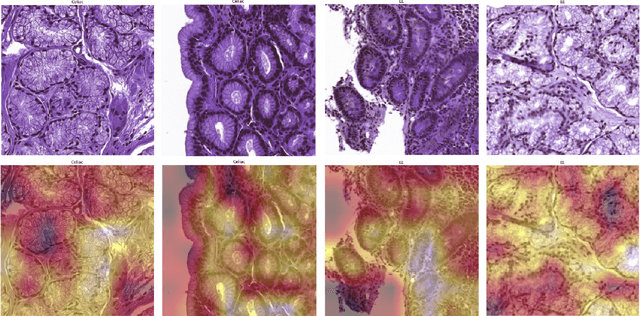
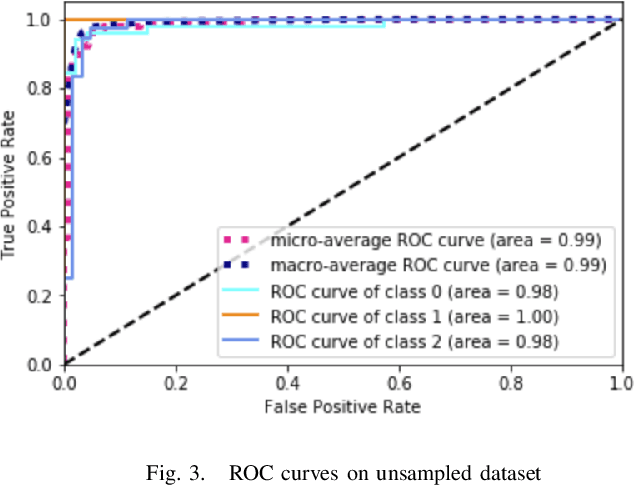
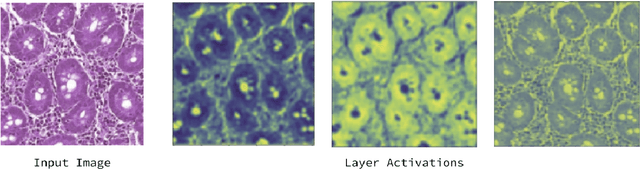
Abstract:Physicians use biopsies to distinguish between different but histologically similar enteropathies. The range of syndromes and pathologies that could cause different gastrointestinal conditions makes this a difficult problem. Recently, deep learning has been used successfully in helping diagnose cancerous tissues in histopathological images. These successes motivated the research presented in this paper, which describes a deep learning approach that distinguishes between Celiac Disease (CD) and Environmental Enteropathy (EE) and normal tissue from digitized duodenal biopsies. Experimental results show accuracies of over 90% for this approach. We also look into interpreting the neural network model using Gradient-weighted Class Activation Mappings and filter activations on input images to understand the visual explanations for the decisions made by the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge