Saurav Sengupta
Detecting Spike Wave Discharges (SWD) using 1-dimensional Residual UNet
Jan 01, 2026Abstract:The manual labeling of events in electroencephalography (EEG) records is time-consuming. This is especially true when EEG recordings are taken continuously over weeks to months. Therefore, a method to automatically label pertinent EEG events reduces the manual workload. Spike wave discharges (SWD), which are the electrographic hallmark of absence seizures, are EEG events that are often labeled manually. While some previous studies have utilized machine learning to automatically segment and classify EEG signals like SWDs, they can be improved. Here we compare the performance of 14 machine learning classifiers on our own manually annotated dataset of 961 hours of EEG recordings from C3H/HeJ mice, including 22,637 labeled SWDs. We find that a 1D UNet performs best for labeling SWDs in this dataset. We also improve the 1D UNet by augmenting our training data and determine that scaling showed the greatest benefit of all augmentation procedures applied. We then compare the 1D UNet with data augmentation, AugUNet1D, against a recently published time- and frequency-based algorithmic approach called "Twin Peaks". AugUNet1D showed superior performance and detected events with more similar features to the SWDs labeled manually. AugUNet1D, pretrained on our manually annotated data or untrained, is made public for others users.
Examining Vision Language Models through Multi-dimensional Experiments with Vision and Text Features
Sep 10, 2025Abstract:Recent research on Vision Language Models (VLMs) suggests that they rely on inherent biases learned during training to respond to questions about visual properties of an image. These biases are exacerbated when VLMs are asked highly specific questions that require focusing on specific areas of the image. For example, a VLM tasked with counting stars on a modified American flag (e.g., with more than 50 stars) will often disregard the visual evidence and fail to answer accurately. We build upon this research and develop a multi-dimensional examination framework to systematically determine which characteristics of the input data, including both the image and the accompanying prompt, lead to such differences in performance. Using open-source VLMs, we further examine how attention values fluctuate with varying input parameters (e.g., image size, number of objects in the image, background color, prompt specificity). This research aims to learn how the behavior of vision language models changes and to explore methods for characterizing such changes. Our results suggest, among other things, that even minor modifications in image characteristics and prompt specificity can lead to large changes in how a VLM formulates its answer and, subsequently, its overall performance.
Towards Robust Multimodal Representation: A Unified Approach with Adaptive Experts and Alignment
Mar 12, 2025Abstract:Healthcare relies on multiple types of data, such as medical images, genetic information, and clinical records, to improve diagnosis and treatment. However, missing data is a common challenge due to privacy restrictions, cost, and technical issues, making many existing multi-modal models unreliable. To address this, we propose a new multi-model model called Mixture of Experts, Symmetric Aligning, and Reconstruction (MoSARe), a deep learning framework that handles incomplete multimodal data while maintaining high accuracy. MoSARe integrates expert selection, cross-modal attention, and contrastive learning to improve feature representation and decision-making. Our results show that MoSARe outperforms existing models in situations when the data is complete. Furthermore, it provides reliable predictions even when some data are missing. This makes it especially useful in real-world healthcare settings, including resource-limited environments. Our code is publicly available at https://github.com/NazaninMn/MoSARe.
Automatic Report Generation for Histopathology images using pre-trained Vision Transformers and BERT
Dec 03, 2023Abstract:Deep learning for histopathology has been successfully used for disease classification, image segmentation and more. However, combining image and text modalities using current state-of-the-art methods has been a challenge due to the high resolution of histopathology images. Automatic report generation for histopathology images is one such challenge. In this work, we show that using an existing pre-trained Vision Transformer in a two-step process of first using it to encode 4096x4096 sized patches of the Whole Slide Image (WSI) and then using it as the encoder and a pre-trained Bidirectional Encoder Representations from Transformers (BERT) model for language modeling-based decoder for report generation, we can build a fairly performant and portable report generation mechanism that takes into account the whole of the high resolution image, instead of just the patches. Our method allows us to not only generate and evaluate captions that describe the image, but also helps us classify the image into tissue types and the gender of the patient as well. Our best performing model achieves a 79.98% accuracy in Tissue Type classification and 66.36% accuracy in classifying the sex of the patient the tissue came from, with a BLEU-4 score of 0.5818 in our caption generation task.
Automatic Report Generation for Histopathology images using pre-trained Vision Transformers
Nov 13, 2023



Abstract:Deep learning for histopathology has been successfully used for disease classification, image segmentation and more. However, combining image and text modalities using current state-of-the-art methods has been a challenge due to the high resolution of histopathology images. Automatic report generation for histopathology images is one such challenge. In this work, we show that using an existing pre-trained Vision Transformer in a two-step process of first using it to encode 4096x4096 sized patches of the Whole Slide Image (WSI) and then using it as the encoder and an LSTM decoder for report generation, we can build a fairly performant and portable report generation mechanism that takes into account the whole of the high resolution image, instead of just the patches. We are also able to use representations from an existing powerful pre-trained hierarchical vision transformer and show its usefulness in not just zero shot classification but also for report generation.
Analyzing historical diagnosis code data from NIH N3C and RECOVER Programs using deep learning to determine risk factors for Long Covid
Oct 05, 2022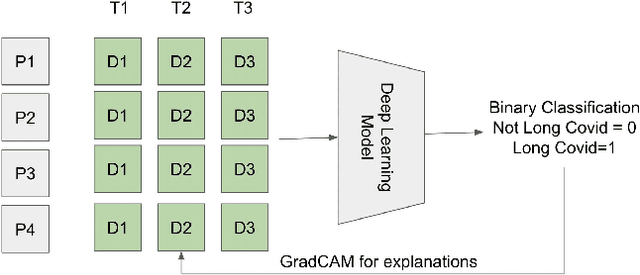
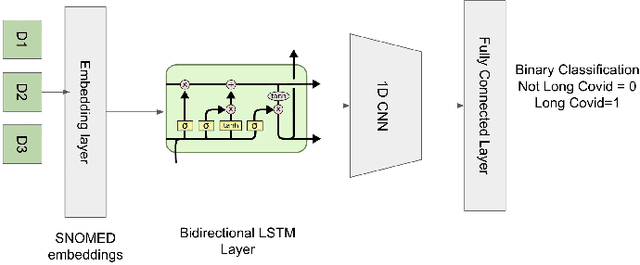
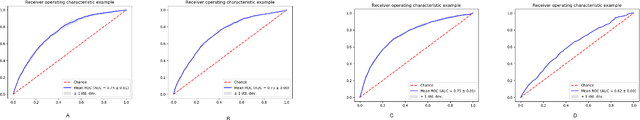
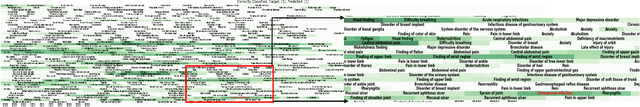
Abstract:Post-acute sequelae of SARS-CoV-2 infection (PASC) or Long COVID is an emerging medical condition that has been observed in several patients with a positive diagnosis for COVID-19. Historical Electronic Health Records (EHR) like diagnosis codes, lab results and clinical notes have been analyzed using deep learning and have been used to predict future clinical events. In this paper, we propose an interpretable deep learning approach to analyze historical diagnosis code data from the National COVID Cohort Collective (N3C) to find the risk factors contributing to developing Long COVID. Using our deep learning approach, we are able to predict if a patient is suffering from Long COVID from a temporally ordered list of diagnosis codes up to 45 days post the first COVID positive test or diagnosis for each patient, with an accuracy of 70.48\%. We are then able to examine the trained model using Gradient-weighted Class Activation Mapping (GradCAM) to give each input diagnoses a score. The highest scored diagnosis were deemed to be the most important for making the correct prediction for a patient. We also propose a way to summarize these top diagnoses for each patient in our cohort and look at their temporal trends to determine which codes contribute towards a positive Long COVID diagnosis.
Deep Learning for Visual Recognition of Environmental Enteropathy and Celiac Disease
Aug 08, 2019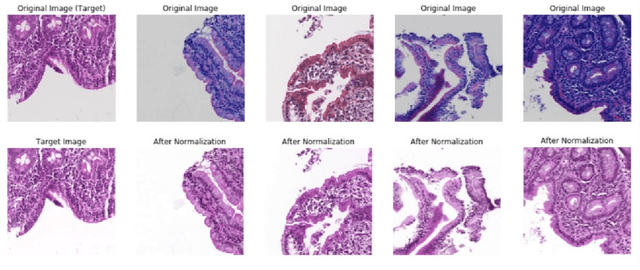
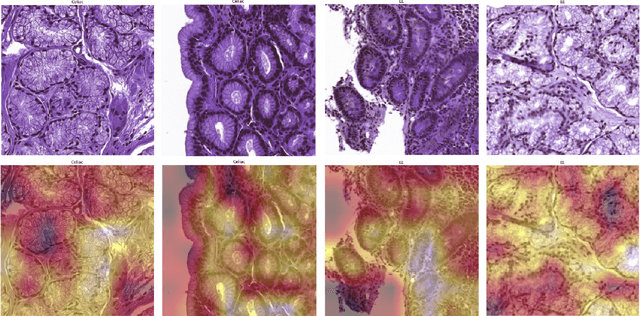
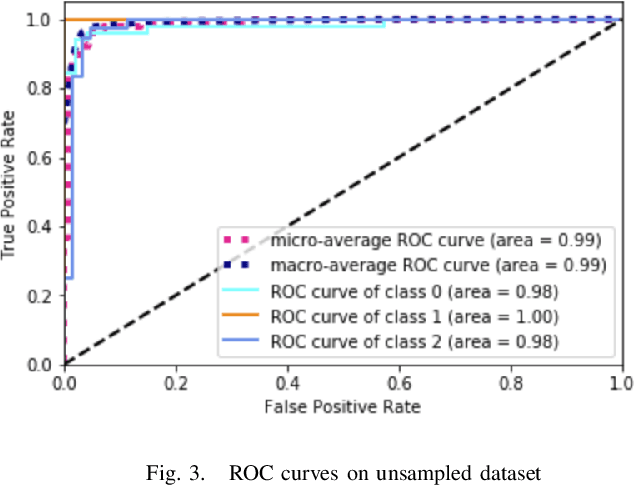
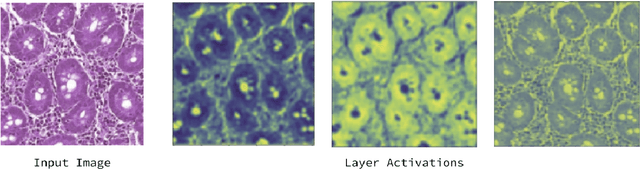
Abstract:Physicians use biopsies to distinguish between different but histologically similar enteropathies. The range of syndromes and pathologies that could cause different gastrointestinal conditions makes this a difficult problem. Recently, deep learning has been used successfully in helping diagnose cancerous tissues in histopathological images. These successes motivated the research presented in this paper, which describes a deep learning approach that distinguishes between Celiac Disease (CD) and Environmental Enteropathy (EE) and normal tissue from digitized duodenal biopsies. Experimental results show accuracies of over 90% for this approach. We also look into interpreting the neural network model using Gradient-weighted Class Activation Mappings and filter activations on input images to understand the visual explanations for the decisions made by the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge