Marium Khan
CeliacNet: Celiac Disease Severity Diagnosis on Duodenal Histopathological Images Using Deep Residual Networks
Oct 07, 2019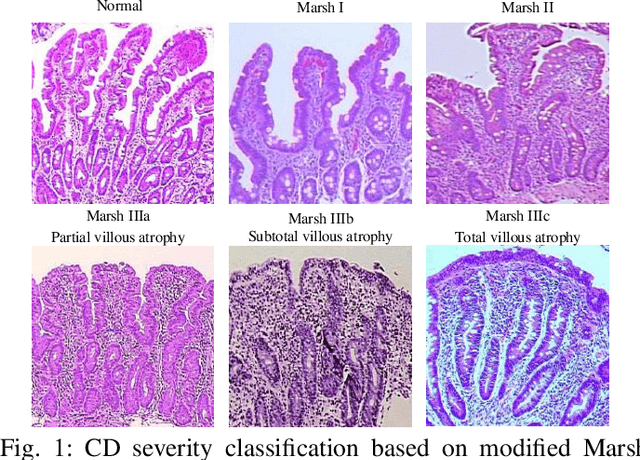
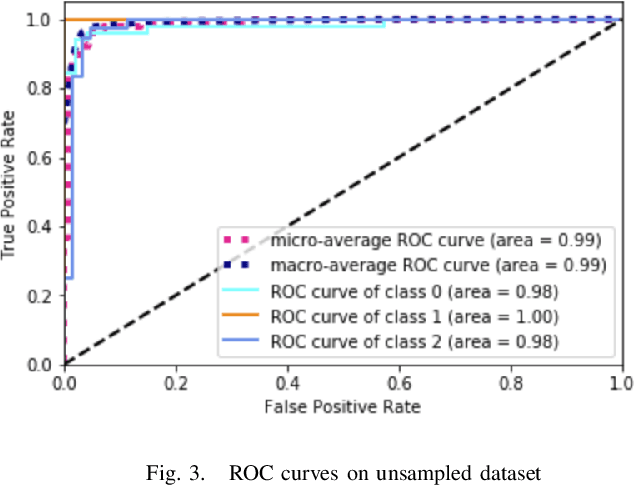
Abstract:Celiac Disease (CD) is a chronic autoimmune disease that affects the small intestine in genetically predisposed children and adults. Gluten exposure triggers an inflammatory cascade which leads to compromised intestinal barrier function. If this enteropathy is unrecognized, this can lead to anemia, decreased bone density, and, in longstanding cases, intestinal cancer. The prevalence of the disorder is 1% in the United States. An intestinal (duodenal) biopsy is considered the "gold standard" for diagnosis. The mild CD might go unnoticed due to non-specific clinical symptoms or mild histologic features. In our current work, we trained a model based on deep residual networks to diagnose CD severity using a histological scoring system called the modified Marsh score. The proposed model was evaluated using an independent set of 120 whole slide images from 15 CD patients and achieved an AUC greater than 0.96 in all classes. These results demonstrate the diagnostic power of the proposed model for CD severity classification using histological images.
Deep Learning for Visual Recognition of Environmental Enteropathy and Celiac Disease
Aug 08, 2019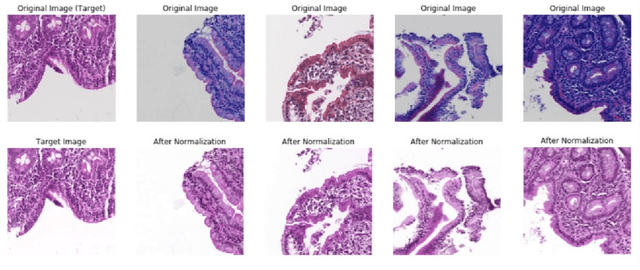
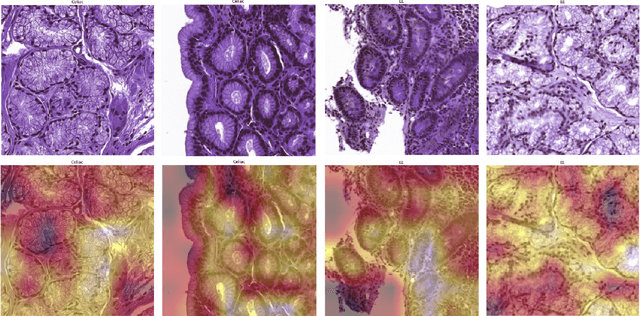
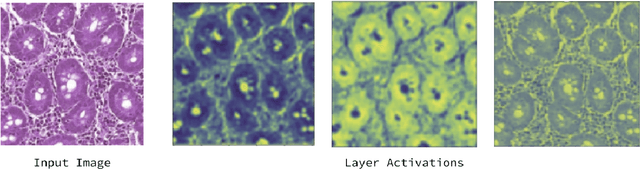
Abstract:Physicians use biopsies to distinguish between different but histologically similar enteropathies. The range of syndromes and pathologies that could cause different gastrointestinal conditions makes this a difficult problem. Recently, deep learning has been used successfully in helping diagnose cancerous tissues in histopathological images. These successes motivated the research presented in this paper, which describes a deep learning approach that distinguishes between Celiac Disease (CD) and Environmental Enteropathy (EE) and normal tissue from digitized duodenal biopsies. Experimental results show accuracies of over 90% for this approach. We also look into interpreting the neural network model using Gradient-weighted Class Activation Mappings and filter activations on input images to understand the visual explanations for the decisions made by the model.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge