Melissa A Haendel
The Cell Ontology in the age of single-cell omics
Jun 10, 2025Abstract:Single-cell omics technologies have transformed our understanding of cellular diversity by enabling high-resolution profiling of individual cells. However, the unprecedented scale and heterogeneity of these datasets demand robust frameworks for data integration and annotation. The Cell Ontology (CL) has emerged as a pivotal resource for achieving FAIR (Findable, Accessible, Interoperable, and Reusable) data principles by providing standardized, species-agnostic terms for canonical cell types - forming a core component of a wide range of platforms and tools. In this paper, we describe the wide variety of uses of CL in these platforms and tools and detail ongoing work to improve and extend CL content including the addition of transcriptomically defined types, working closely with major atlasing efforts including the Human Cell Atlas and the Brain Initiative Cell Atlas Network to support their needs. We cover the challenges and future plans for harmonising classical and transcriptomic cell type definitions, integrating markers and using Large Language Models (LLMs) to improve content and efficiency of CL workflows.
CurateGPT: A flexible language-model assisted biocuration tool
Oct 29, 2024



Abstract:Effective data-driven biomedical discovery requires data curation: a time-consuming process of finding, organizing, distilling, integrating, interpreting, annotating, and validating diverse information into a structured form suitable for databases and knowledge bases. Accurate and efficient curation of these digital assets is critical to ensuring that they are FAIR, trustworthy, and sustainable. Unfortunately, expert curators face significant time and resource constraints. The rapid pace of new information being published daily is exceeding their capacity for curation. Generative AI, exemplified by instruction-tuned large language models (LLMs), has opened up new possibilities for assisting human-driven curation. The design philosophy of agents combines the emerging abilities of generative AI with more precise methods. A curator's tasks can be aided by agents for performing reasoning, searching ontologies, and integrating knowledge across external sources, all efforts otherwise requiring extensive manual effort. Our LLM-driven annotation tool, CurateGPT, melds the power of generative AI together with trusted knowledge bases and literature sources. CurateGPT streamlines the curation process, enhancing collaboration and efficiency in common workflows. Compared to direct interaction with an LLM, CurateGPT's agents enable access to information beyond that in the LLM's training data and they provide direct links to the data supporting each claim. This helps curators, researchers, and engineers scale up curation efforts to keep pace with the ever-increasing volume of scientific data.
Dynamic Retrieval Augmented Generation of Ontologies using Artificial Intelligence (DRAGON-AI)
Dec 18, 2023Abstract:Ontologies are fundamental components of informatics infrastructure in domains such as biomedical, environmental, and food sciences, representing consensus knowledge in an accurate and computable form. However, their construction and maintenance demand substantial resources, necessitating substantial collaborative efforts of domain experts, curators, and ontology experts. We present Dynamic Retrieval Augmented Generation of Ontologies using AI (DRAGON-AI), an ontology generation method employing Large Language Models (LLMs) and Retrieval Augmented Generation (RAG). This method can generate textual and logical ontology components, drawing from existing knowledge in multiple ontologies, as well as unstructured textual sources. We assessed DRAGON-AI across ten diverse ontologies, making use of extensive manual evaluation of results. We demonstrate high precision for relationship generation, close to but lower than precision from logic-based reasoning. We also demonstrate definition generation comparable with but lower than human-generated definitions. Notably, expert evaluators were better able to discern subtle flaws in AI-generated definitions. We also demonstrated the ability of DRAGON-AI to incorporate natural language instructions in the form of GitHub issues. These findings suggest DRAGON-AI's potential to substantially aid the manual ontology construction process. However, our results also underscore the importance of having expert curators and ontology editors drive the ontology generation process.
MapperGPT: Large Language Models for Linking and Mapping Entities
Oct 05, 2023Abstract:Aligning terminological resources, including ontologies, controlled vocabularies, taxonomies, and value sets is a critical part of data integration in many domains such as healthcare, chemistry, and biomedical research. Entity mapping is the process of determining correspondences between entities across these resources, such as gene identifiers, disease concepts, or chemical entity identifiers. Many tools have been developed to compute such mappings based on common structural features and lexical information such as labels and synonyms. Lexical approaches in particular often provide very high recall, but low precision, due to lexical ambiguity. As a consequence of this, mapping efforts often resort to a labor intensive manual mapping refinement through a human curator. Large Language Models (LLMs), such as the ones employed by ChatGPT, have generalizable abilities to perform a wide range of tasks, including question-answering and information extraction. Here we present MapperGPT, an approach that uses LLMs to review and refine mapping relationships as a post-processing step, in concert with existing high-recall methods that are based on lexical and structural heuristics. We evaluated MapperGPT on a series of alignment tasks from different domains, including anatomy, developmental biology, and renal diseases. We devised a collection of tasks that are designed to be particularly challenging for lexical methods. We show that when used in combination with high-recall methods, MapperGPT can provide a substantial improvement in accuracy, beating state-of-the-art (SOTA) methods such as LogMap.
An evaluation of GPT models for phenotype concept recognition
Sep 29, 2023Abstract:Objective: Clinical deep phenotyping plays a critical role in both the diagnosis of patients with rare disorders as well as in building care coordination plans. The process relies on modelling and curating patient profiles using ontology concepts, usually from the Human Phenotype Ontology. Machine learning methods have been widely adopted to support this phenotype concept recognition task. With the significant shift in the use of large language models (LLMs) for most NLP tasks, herewithin, we examine the performance of the latest Generative Pre-trained Transformer (GPT) models underpinning ChatGPT in clinical deep phenotyping. Materials and Methods: The experimental setup of the study included seven prompts of various levels of specificity, two GPT models (gpt-3.5 and gpt-4.0) and an established gold standard for phenotype recognition. Results: Our results show that, currently, these models have not yet achieved state of the art performance. The best run, using few-shots learning, achieved 0.41 F1 score, compared to a 0.62 F1 score achieved by the current best in class tool. Conclusion: The non-deterministic nature of the outcomes and the lack of concordance between different runs using the same prompt and input makes the use of these LLMs in clinical settings problematic.
Analyzing historical diagnosis code data from NIH N3C and RECOVER Programs using deep learning to determine risk factors for Long Covid
Oct 05, 2022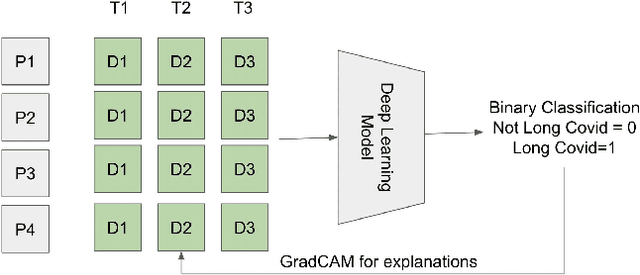
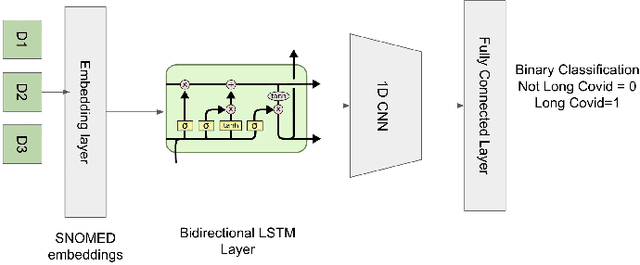
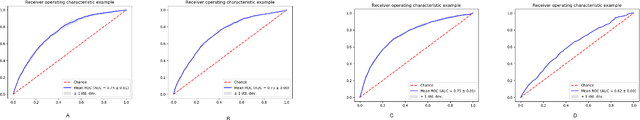
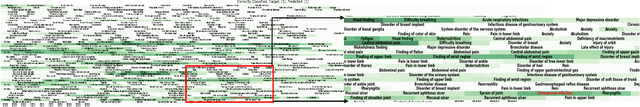
Abstract:Post-acute sequelae of SARS-CoV-2 infection (PASC) or Long COVID is an emerging medical condition that has been observed in several patients with a positive diagnosis for COVID-19. Historical Electronic Health Records (EHR) like diagnosis codes, lab results and clinical notes have been analyzed using deep learning and have been used to predict future clinical events. In this paper, we propose an interpretable deep learning approach to analyze historical diagnosis code data from the National COVID Cohort Collective (N3C) to find the risk factors contributing to developing Long COVID. Using our deep learning approach, we are able to predict if a patient is suffering from Long COVID from a temporally ordered list of diagnosis codes up to 45 days post the first COVID positive test or diagnosis for each patient, with an accuracy of 70.48\%. We are then able to examine the trained model using Gradient-weighted Class Activation Mapping (GradCAM) to give each input diagnoses a score. The highest scored diagnosis were deemed to be the most important for making the correct prediction for a patient. We also propose a way to summarize these top diagnoses for each patient in our cohort and look at their temporal trends to determine which codes contribute towards a positive Long COVID diagnosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge