Nicolas Matentzoglu
The Cell Ontology in the age of single-cell omics
Jun 10, 2025Abstract:Single-cell omics technologies have transformed our understanding of cellular diversity by enabling high-resolution profiling of individual cells. However, the unprecedented scale and heterogeneity of these datasets demand robust frameworks for data integration and annotation. The Cell Ontology (CL) has emerged as a pivotal resource for achieving FAIR (Findable, Accessible, Interoperable, and Reusable) data principles by providing standardized, species-agnostic terms for canonical cell types - forming a core component of a wide range of platforms and tools. In this paper, we describe the wide variety of uses of CL in these platforms and tools and detail ongoing work to improve and extend CL content including the addition of transcriptomically defined types, working closely with major atlasing efforts including the Human Cell Atlas and the Brain Initiative Cell Atlas Network to support their needs. We cover the challenges and future plans for harmonising classical and transcriptomic cell type definitions, integrating markers and using Large Language Models (LLMs) to improve content and efficiency of CL workflows.
MapperGPT: Large Language Models for Linking and Mapping Entities
Oct 05, 2023Abstract:Aligning terminological resources, including ontologies, controlled vocabularies, taxonomies, and value sets is a critical part of data integration in many domains such as healthcare, chemistry, and biomedical research. Entity mapping is the process of determining correspondences between entities across these resources, such as gene identifiers, disease concepts, or chemical entity identifiers. Many tools have been developed to compute such mappings based on common structural features and lexical information such as labels and synonyms. Lexical approaches in particular often provide very high recall, but low precision, due to lexical ambiguity. As a consequence of this, mapping efforts often resort to a labor intensive manual mapping refinement through a human curator. Large Language Models (LLMs), such as the ones employed by ChatGPT, have generalizable abilities to perform a wide range of tasks, including question-answering and information extraction. Here we present MapperGPT, an approach that uses LLMs to review and refine mapping relationships as a post-processing step, in concert with existing high-recall methods that are based on lexical and structural heuristics. We evaluated MapperGPT on a series of alignment tasks from different domains, including anatomy, developmental biology, and renal diseases. We devised a collection of tasks that are designed to be particularly challenging for lexical methods. We show that when used in combination with high-recall methods, MapperGPT can provide a substantial improvement in accuracy, beating state-of-the-art (SOTA) methods such as LogMap.
Structured prompt interrogation and recursive extraction of semantics (SPIRES): A method for populating knowledge bases using zero-shot learning
Apr 05, 2023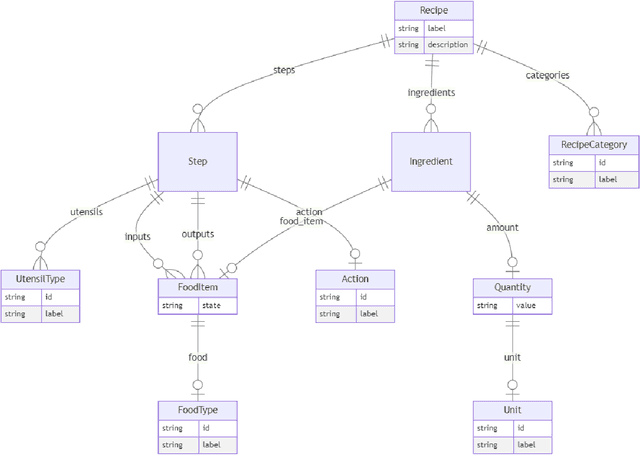
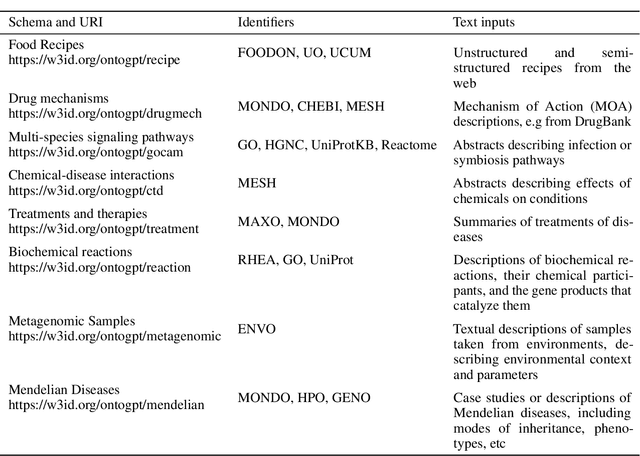
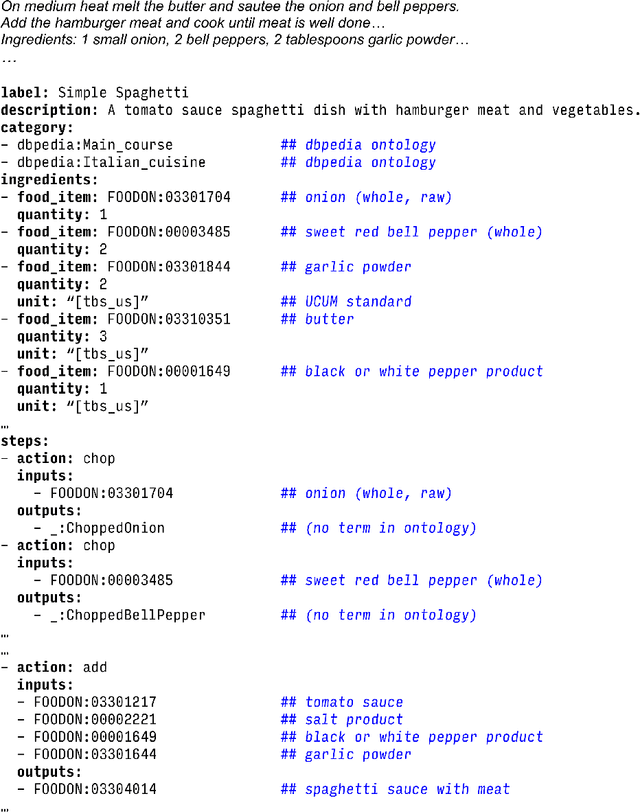
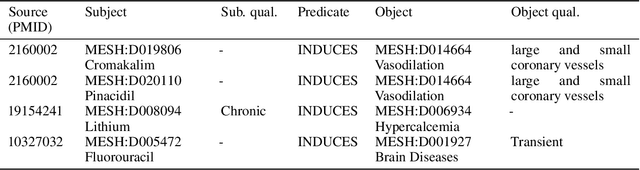
Abstract:Creating knowledge bases and ontologies is a time consuming task that relies on a manual curation. AI/NLP approaches can assist expert curators in populating these knowledge bases, but current approaches rely on extensive training data, and are not able to populate arbitrary complex nested knowledge schemas. Here we present Structured Prompt Interrogation and Recursive Extraction of Semantics (SPIRES), a Knowledge Extraction approach that relies on the ability of Large Language Models (LLMs) to perform zero-shot learning (ZSL) and general-purpose query answering from flexible prompts and return information conforming to a specified schema. Given a detailed, user-defined knowledge schema and an input text, SPIRES recursively performs prompt interrogation against GPT-3+ to obtain a set of responses matching the provided schema. SPIRES uses existing ontologies and vocabularies to provide identifiers for all matched elements. We present examples of use of SPIRES in different domains, including extraction of food recipes, multi-species cellular signaling pathways, disease treatments, multi-step drug mechanisms, and chemical to disease causation graphs. Current SPIRES accuracy is comparable to the mid-range of existing Relation Extraction (RE) methods, but has the advantage of easy customization, flexibility, and, crucially, the ability to perform new tasks in the absence of any training data. This method supports a general strategy of leveraging the language interpreting capabilities of LLMs to assemble knowledge bases, assisting manual knowledge curation and acquisition while supporting validation with publicly-available databases and ontologies external to the LLM. SPIRES is available as part of the open source OntoGPT package: https://github.com/ monarch-initiative/ontogpt.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge