Vincent Emonet
SPARQL-LLM: Real-Time SPARQL Query Generation from Natural Language Questions
Dec 16, 2025Abstract:The advent of large language models is contributing to the emergence of novel approaches that promise to better tackle the challenge of generating structured queries, such as SPARQL queries, from natural language. However, these new approaches mostly focus on response accuracy over a single source while ignoring other evaluation criteria, such as federated query capability over distributed data stores, as well as runtime and cost to generate SPARQL queries. Consequently, they are often not production-ready or easy to deploy over (potentially federated) knowledge graphs with good accuracy. To mitigate these issues, in this paper, we extend our previous work and describe and systematically evaluate SPARQL-LLM, an open-source and triplestore-agnostic approach, powered by lightweight metadata, that generates SPARQL queries from natural language text. First, we describe its architecture, which consists of dedicated components for metadata indexing, prompt building, and query generation and execution. Then, we evaluate it based on a state-of-the-art challenge with multilingual questions, and a collection of questions from three of the most prevalent knowledge graphs within the field of bioinformatics. Our results demonstrate a substantial increase of 24% in the F1 Score on the state-of-the-art challenge, adaptability to high-resource languages such as English and Spanish, as well as ability to form complex and federated bioinformatics queries. Furthermore, we show that SPARQL-LLM is up to 36x faster than other systems participating in the challenge, while costing a maximum of $0.01 per question, making it suitable for real-time, low-cost text-to-SPARQL applications. One such application deployed over real-world decentralized knowledge graphs can be found at https://www.expasy.org/chat.
LLM-based SPARQL Query Generation from Natural Language over Federated Knowledge Graphs
Oct 10, 2024Abstract:We introduce a Retrieval-Augmented Generation (RAG) system for translating user questions into accurate federated SPARQL queries over bioinformatics knowledge graphs (KGs) leveraging Large Language Models (LLMs). To enhance accuracy and reduce hallucinations in query generation, our system utilises metadata from the KGs, including query examples and schema information, and incorporates a validation step to correct generated queries. The system is available online at chat.expasy.org.
A large collection of bioinformatics question-query pairs over federated knowledge graphs: methodology and applications
Oct 08, 2024



Abstract:Background. In the last decades, several life science resources have structured data using the same framework and made these accessible using the same query language to facilitate interoperability. Knowledge graphs have seen increased adoption in bioinformatics due to their advantages for representing data in a generic graph format. For example, yummydata.org catalogs more than 60 knowledge graphs accessible through SPARQL, a technical query language. Although SPARQL allows powerful, expressive queries, even across physically distributed knowledge graphs, formulating such queries is a challenge for most users. Therefore, to guide users in retrieving the relevant data, many of these resources provide representative examples. These examples can also be an important source of information for machine learning, if a sufficiently large number of examples are provided and published in a common, machine-readable and standardized format across different resources. Findings. We introduce a large collection of human-written natural language questions and their corresponding SPARQL queries over federated bioinformatics knowledge graphs (KGs) collected for several years across different research groups at the SIB Swiss Institute of Bioinformatics. The collection comprises more than 1000 example questions and queries, including 65 federated queries. We propose a methodology to uniformly represent the examples with minimal metadata, based on existing standards. Furthermore, we introduce an extensive set of open-source applications, including query graph visualizations and smart query editors, easily reusable by KG maintainers who adopt the proposed methodology. Conclusions. We encourage the community to adopt and extend the proposed methodology, towards richer KG metadata and improved Semantic Web services.
Structured prompt interrogation and recursive extraction of semantics (SPIRES): A method for populating knowledge bases using zero-shot learning
Apr 05, 2023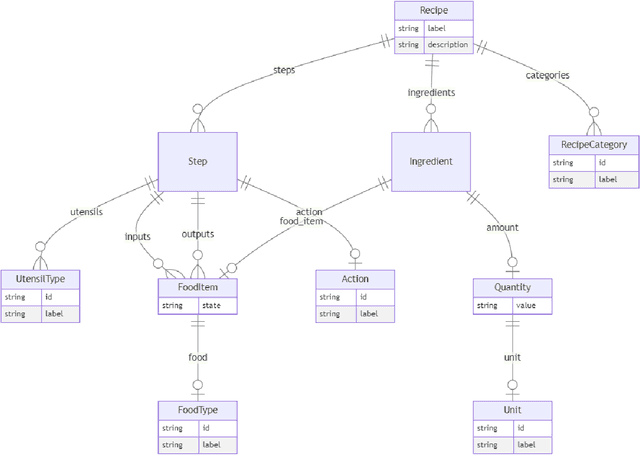
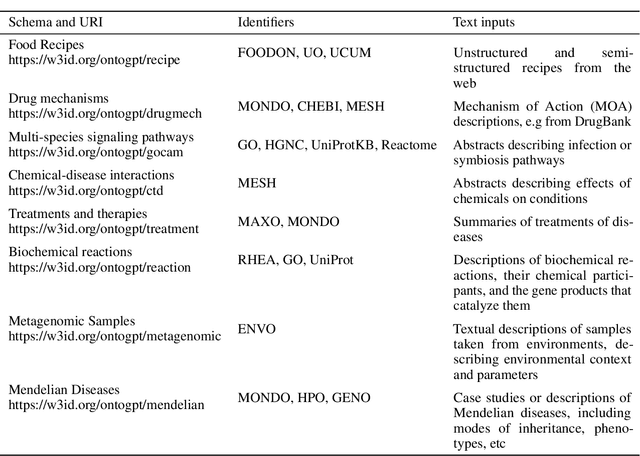
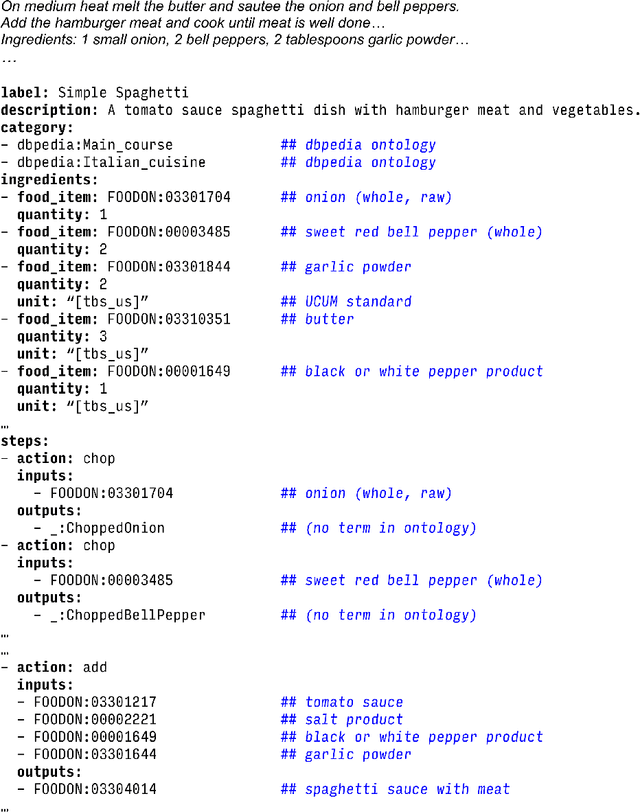
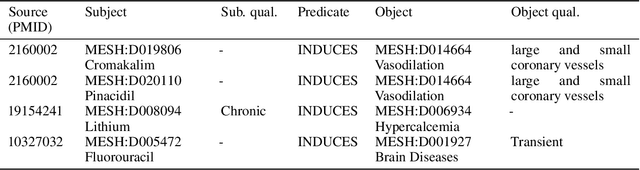
Abstract:Creating knowledge bases and ontologies is a time consuming task that relies on a manual curation. AI/NLP approaches can assist expert curators in populating these knowledge bases, but current approaches rely on extensive training data, and are not able to populate arbitrary complex nested knowledge schemas. Here we present Structured Prompt Interrogation and Recursive Extraction of Semantics (SPIRES), a Knowledge Extraction approach that relies on the ability of Large Language Models (LLMs) to perform zero-shot learning (ZSL) and general-purpose query answering from flexible prompts and return information conforming to a specified schema. Given a detailed, user-defined knowledge schema and an input text, SPIRES recursively performs prompt interrogation against GPT-3+ to obtain a set of responses matching the provided schema. SPIRES uses existing ontologies and vocabularies to provide identifiers for all matched elements. We present examples of use of SPIRES in different domains, including extraction of food recipes, multi-species cellular signaling pathways, disease treatments, multi-step drug mechanisms, and chemical to disease causation graphs. Current SPIRES accuracy is comparable to the mid-range of existing Relation Extraction (RE) methods, but has the advantage of easy customization, flexibility, and, crucially, the ability to perform new tasks in the absence of any training data. This method supports a general strategy of leveraging the language interpreting capabilities of LLMs to assemble knowledge bases, assisting manual knowledge curation and acquisition while supporting validation with publicly-available databases and ontologies external to the LLM. SPIRES is available as part of the open source OntoGPT package: https://github.com/ monarch-initiative/ontogpt.
Knowledge Graphs Evolution and Preservation -- A Technical Report from ISWS 2019
Dec 22, 2020
Abstract:One of the grand challenges discussed during the Dagstuhl Seminar "Knowledge Graphs: New Directions for Knowledge Representation on the Semantic Web" and described in its report is that of a: "Public FAIR Knowledge Graph of Everything: We increasingly see the creation of knowledge graphs that capture information about the entirety of a class of entities. [...] This grand challenge extends this further by asking if we can create a knowledge graph of "everything" ranging from common sense concepts to location based entities. This knowledge graph should be "open to the public" in a FAIR manner democratizing this mass amount of knowledge." Although linked open data (LOD) is one knowledge graph, it is the closest realisation (and probably the only one) to a public FAIR Knowledge Graph (KG) of everything. Surely, LOD provides a unique testbed for experimenting and evaluating research hypotheses on open and FAIR KG. One of the most neglected FAIR issues about KGs is their ongoing evolution and long term preservation. We want to investigate this problem, that is to understand what preserving and supporting the evolution of KGs means and how these problems can be addressed. Clearly, the problem can be approached from different perspectives and may require the development of different approaches, including new theories, ontologies, metrics, strategies, procedures, etc. This document reports a collaborative effort performed by 9 teams of students, each guided by a senior researcher as their mentor, attending the International Semantic Web Research School (ISWS 2019). Each team provides a different perspective to the problem of knowledge graph evolution substantiated by a set of research questions as the main subject of their investigation. In addition, they provide their working definition for KG preservation and evolution.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge