P. Thomas Fletcher
Electrical & Computer Engineering, University of Virginia
Point-Based Shape Representation Generation with a Correspondence-Preserving Diffusion Model
Aug 05, 2025Abstract:We propose a diffusion model designed to generate point-based shape representations with correspondences. Traditional statistical shape models have considered point correspondences extensively, but current deep learning methods do not take them into account, focusing on unordered point clouds instead. Current deep generative models for point clouds do not address generating shapes with point correspondences between generated shapes. This work aims to formulate a diffusion model that is capable of generating realistic point-based shape representations, which preserve point correspondences that are present in the training data. Using shape representation data with correspondences derived from Open Access Series of Imaging Studies 3 (OASIS-3), we demonstrate that our correspondence-preserving model effectively generates point-based hippocampal shape representations that are highly realistic compared to existing methods. We further demonstrate the applications of our generative model by downstream tasks, such as conditional generation of healthy and AD subjects and predicting morphological changes of disease progression by counterfactual generation.
MedIL: Implicit Latent Spaces for Generating Heterogeneous Medical Images at Arbitrary Resolutions
Apr 12, 2025



Abstract:In this work, we introduce MedIL, a first-of-its-kind autoencoder built for encoding medical images with heterogeneous sizes and resolutions for image generation. Medical images are often large and heterogeneous, where fine details are of vital clinical importance. Image properties change drastically when considering acquisition equipment, patient demographics, and pathology, making realistic medical image generation challenging. Recent work in latent diffusion models (LDMs) has shown success in generating images resampled to a fixed-size. However, this is a narrow subset of the resolutions native to image acquisition, and resampling discards fine anatomical details. MedIL utilizes implicit neural representations to treat images as continuous signals, where encoding and decoding can be performed at arbitrary resolutions without prior resampling. We quantitatively and qualitatively show how MedIL compresses and preserves clinically-relevant features over large multi-site, multi-resolution datasets of both T1w brain MRIs and lung CTs. We further demonstrate how MedIL can influence the quality of images generated with a diffusion model, and discuss how MedIL can enhance generative models to resemble raw clinical acquisitions.
Measuring Feature Dependency of Neural Networks by Collapsing Feature Dimensions in the Data Manifold
Apr 18, 2024



Abstract:This paper introduces a new technique to measure the feature dependency of neural network models. The motivation is to better understand a model by querying whether it is using information from human-understandable features, e.g., anatomical shape, volume, or image texture. Our method is based on the principle that if a model is dependent on a feature, then removal of that feature should significantly harm its performance. A targeted feature is "removed" by collapsing the dimension in the data distribution that corresponds to that feature. We perform this by moving data points along the feature dimension to a baseline feature value while staying on the data manifold, as estimated by a deep generative model. Then we observe how the model's performance changes on the modified test data set, with the target feature dimension removed. We test our method on deep neural network models trained on synthetic image data with known ground truth, an Alzheimer's disease prediction task using MRI and hippocampus segmentations from the OASIS-3 dataset, and a cell nuclei classification task using the Lizard dataset.
Learning Spatially-Continuous Fiber Orientation Functions
Dec 10, 2023Abstract:Our understanding of the human connectome is fundamentally limited by the resolution of diffusion MR images. Reconstructing a connectome's constituent neural pathways with tractography requires following a continuous field of fiber directions. Typically, this field is found with simple trilinear interpolation in low-resolution, noisy diffusion MRIs. However, trilinear interpolation struggles following fine-scale changes in low-quality data. Recent deep learning methods in super-resolving diffusion MRIs have focused on upsampling to a fixed spatial grid, but this does not satisfy tractography's need for a continuous field. In this work, we propose FENRI, a novel method that learns spatially-continuous fiber orientation density functions from low-resolution diffusion-weighted images. To quantify FENRI's capabilities in tractography, we also introduce an expanded simulated dataset built for evaluating deep-learning tractography models. We demonstrate that FENRI accurately predicts high-resolution fiber orientations from realistic low-quality data, and that FENRI-based tractography offers improved streamline reconstruction over the current use of trilinear interpolation.
Quantifying Hippocampal Shape Asymmetry in Alzheimer's Disease Using Optimal Shape Correspondences
Dec 02, 2023Abstract:Hippocampal atrophy in Alzheimer's disease (AD) is asymmetric and spatially inhomogeneous. While extensive work has been done on volume and shape analysis of atrophy of the hippocampus in AD, less attention has been given to hippocampal asymmetry specifically. Previous studies of hippocampal asymmetry are limited to global volume or shape measures, which don't localize shape asymmetry at the point level. In this paper, we propose to quantify localized shape asymmetry by optimizing point correspondences between left and right hippocampi within a subject, while simultaneously favoring a compact statistical shape model of the entire sample. To account for related variables that have impact on AD and healthy subject differences, we build linear models with other confounding factors. Our results on the OASIS3 dataset demonstrate that compared to using volumetric information, shape asymmetry reveals fine-grained, localized differences that indicate the hippocampal regions of most significant shape asymmetry in AD patients.
Feature Gradient Flow for Interpreting Deep Neural Networks in Head and Neck Cancer Prediction
Jul 24, 2023Abstract:This paper introduces feature gradient flow, a new technique for interpreting deep learning models in terms of features that are understandable to humans. The gradient flow of a model locally defines nonlinear coordinates in the input data space representing the information the model is using to make its decisions. Our idea is to measure the agreement of interpretable features with the gradient flow of a model. To then evaluate the importance of a particular feature to the model, we compare that feature's gradient flow measure versus that of a baseline noise feature. We then develop a technique for training neural networks to be more interpretable by adding a regularization term to the loss function that encourages the model gradients to align with those of chosen interpretable features. We test our method in a convolutional neural network prediction of distant metastasis of head and neck cancer from a computed tomography dataset from the Cancer Imaging Archive.
NASDM: Nuclei-Aware Semantic Histopathology Image Generation Using Diffusion Models
Mar 20, 2023



Abstract:In recent years, computational pathology has seen tremendous progress driven by deep learning methods in segmentation and classification tasks aiding prognostic and diagnostic settings. Nuclei segmentation, for instance, is an important task for diagnosing different cancers. However, training deep learning models for nuclei segmentation requires large amounts of annotated data, which is expensive to collect and label. This necessitates explorations into generative modeling of histopathological images. In this work, we use recent advances in conditional diffusion modeling to formulate a first-of-its-kind nuclei-aware semantic tissue generation framework (NASDM) which can synthesize realistic tissue samples given a semantic instance mask of up to six different nuclei types, enabling pixel-perfect nuclei localization in generated samples. These synthetic images are useful in applications in pathology pedagogy, validation of models, and supplementation of existing nuclei segmentation datasets. We demonstrate that NASDM is able to synthesize high-quality histopathology images of the colon with superior quality and semantic controllability over existing generative methods.
Deep Learning the Shape of the Brain Connectome
Mar 06, 2022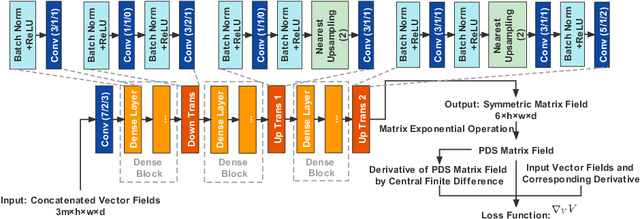
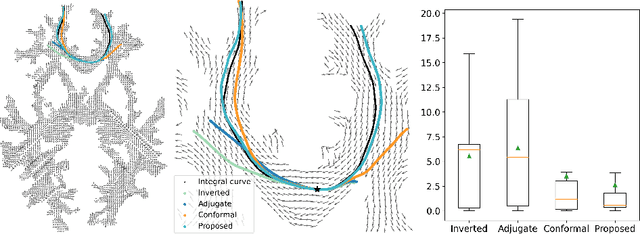
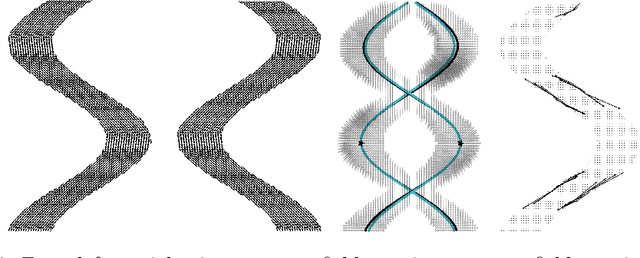
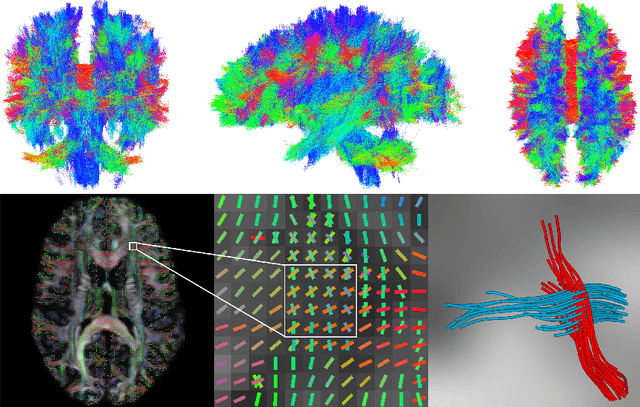
Abstract:To statistically study the variability and differences between normal and abnormal brain connectomes, a mathematical model of the neural connections is required. In this paper, we represent the brain connectome as a Riemannian manifold, which allows us to model neural connections as geodesics. We show for the first time how one can leverage deep neural networks to estimate a Riemannian metric of the brain that can accommodate fiber crossings and is a natural modeling tool to infer the shape of the brain from DWMRI. Our method achieves excellent performance in geodesic-white-matter-pathway alignment and tackles the long-standing issue in previous methods: the inability to recover the crossing fibers with high fidelity.
Integrated Construction of Multimodal Atlases with Structural Connectomes in the Space of Riemannian Metrics
Sep 20, 2021

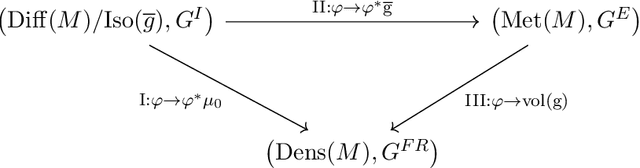

Abstract:The structural network of the brain, or structural connectome, can be represented by fiber bundles generated by a variety of tractography methods. While such methods give qualitative insights into brain structure, there is controversy over whether they can provide quantitative information, especially at the population level. In order to enable population-level statistical analysis of the structural connectome, we propose representing a connectome as a Riemannian metric, which is a point on an infinite-dimensional manifold. We equip this manifold with the Ebin metric, a natural metric structure for this space, to get a Riemannian manifold along with its associated geometric properties. We then use this Riemannian framework to apply object-oriented statistical analysis to define an atlas as the Fr\'echet mean of a population of Riemannian metrics. This formulation ties into the existing framework for diffeomorphic construction of image atlases, allowing us to construct a multimodal atlas by simultaneously integrating complementary white matter structure details from DWMRI and cortical details from T1-weighted MRI. We illustrate our framework with 2D data examples of connectome registration and atlas formation. Finally, we build an example 3D multimodal atlas using T1 images and connectomes derived from diffusion tensors estimated from a subset of subjects from the Human Connectome Project.
Structural Connectome Atlas Construction in the Space of Riemannian Metrics
Mar 09, 2021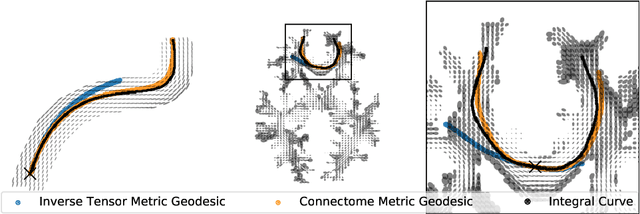
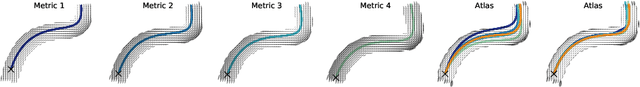
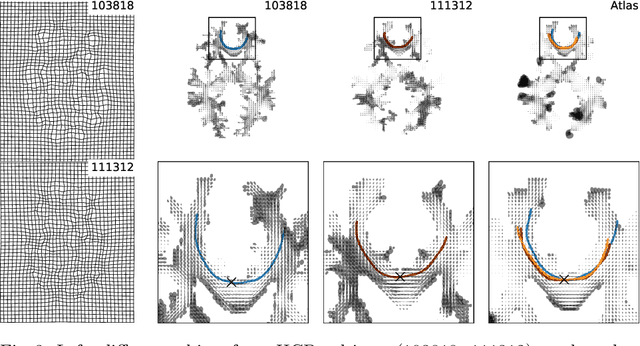
Abstract:The structural connectome is often represented by fiber bundles generated from various types of tractography. We propose a method of analyzing connectomes by representing them as a Riemannian metric, thereby viewing them as points in an infinite-dimensional manifold. After equipping this space with a natural metric structure, the Ebin metric, we apply object-oriented statistical analysis to define an atlas as the Fr\'echet mean of a population of Riemannian metrics. We demonstrate connectome registration and atlas formation using connectomes derived from diffusion tensors estimated from a subset of subjects from the Human Connectome Project.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge