Jaideep Kapur
Quantifying Hippocampal Shape Asymmetry in Alzheimer's Disease Using Optimal Shape Correspondences
Dec 02, 2023Abstract:Hippocampal atrophy in Alzheimer's disease (AD) is asymmetric and spatially inhomogeneous. While extensive work has been done on volume and shape analysis of atrophy of the hippocampus in AD, less attention has been given to hippocampal asymmetry specifically. Previous studies of hippocampal asymmetry are limited to global volume or shape measures, which don't localize shape asymmetry at the point level. In this paper, we propose to quantify localized shape asymmetry by optimizing point correspondences between left and right hippocampi within a subject, while simultaneously favoring a compact statistical shape model of the entire sample. To account for related variables that have impact on AD and healthy subject differences, we build linear models with other confounding factors. Our results on the OASIS3 dataset demonstrate that compared to using volumetric information, shape asymmetry reveals fine-grained, localized differences that indicate the hippocampal regions of most significant shape asymmetry in AD patients.
Cellcounter: a deep learning framework for high-fidelity spatial localization of neurons
Mar 18, 2021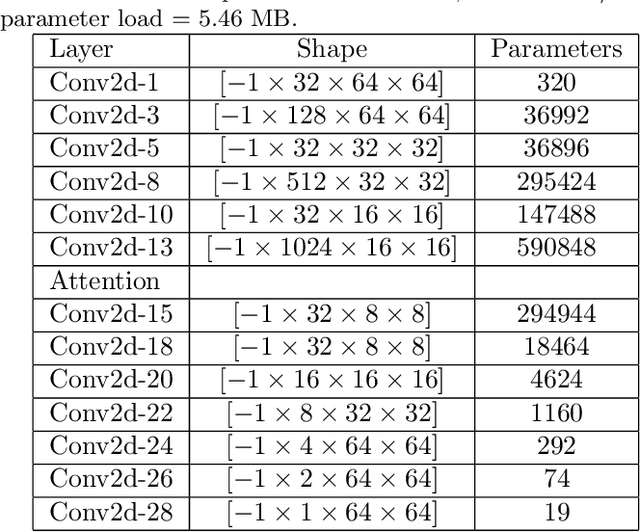
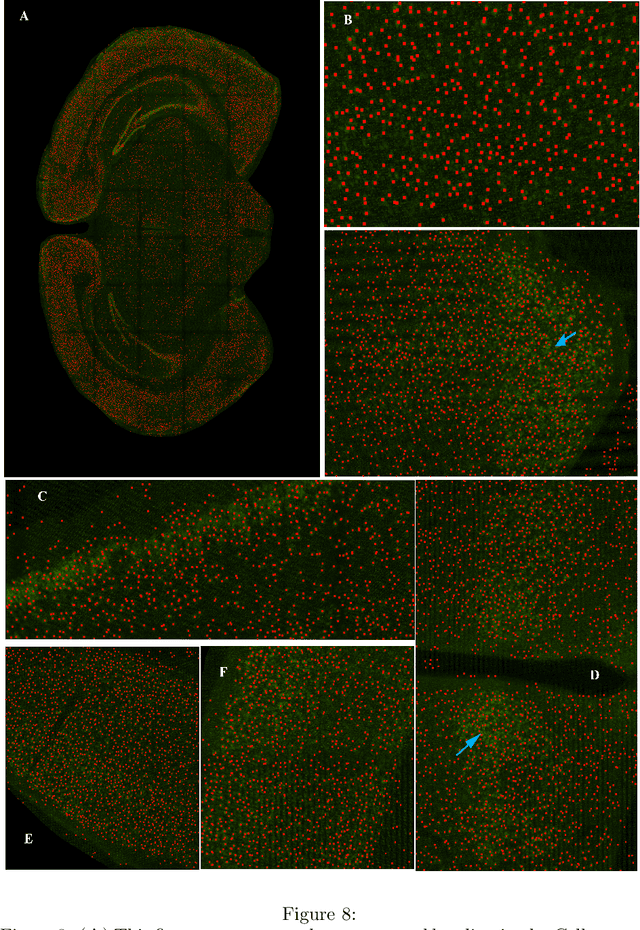
Abstract:Many neuroscientific applications require robust and accurate localization of neurons. It is still an unsolved problem because of the enormous variation in intensity, texture, spatial overlap, morphology and background artifacts. In addition, curation of a large dataset containing complete manual annotation of neurons from high-resolution images to train a classifier requires significant time and effort. We present Cellcounter, a deep learning-based model trained on images containing incompletely-annotated neurons with highly-varied morphology and control images containing artifacts and background structures. Leveraging the striking self-learning ability, Cellcounter gradually labels neurons, obviating the need for time-intensive complete annotation. Cellcounter shows its efficacy over the state of the arts in the accurate localization of neurons while significantly reducing false-positive detection in several protocols.
Enhanced Center Coding for Cell Detection with Convolutional Neural Networks
Apr 18, 2019



Abstract:Cell imaging and analysis are fundamental to biomedical research because cells are the basic functional units of life. Among different cell-related analysis, cell counting and detection are widely used. In this paper, we focus on one common step of learning-based cell counting approaches: coding the raw dot labels into more suitable maps for learning. Two criteria of coding raw dot labels are discussed, and a new coding scheme is proposed in this paper. The two criteria measure how easy it is to train the model with a coding scheme, and how robust the recovered raw dot labels are when predicting. The most compelling advantage of the proposed coding scheme is the ability to distinguish neighboring cells in crowded regions. Cell counting and detection experiments are conducted for five coding schemes on four types of cells and two network architectures. The proposed coding scheme improves the counting accuracy versus the widely-used Gaussian and rectangle kernels up to 12%, and also improves the detection accuracy versus the common proximity coding up to 14%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge